




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (10): 72-83.DOI: 10.13304/j.nykjdb.2024.0297
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Qiujie CHEN( ), Yumei HUANG, Jinyan XIAN, Kaitong LI, Jielan HUANG, Xiaoxiu SHI, Liuping WANG(
), Yumei HUANG, Jinyan XIAN, Kaitong LI, Jielan HUANG, Xiaoxiu SHI, Liuping WANG( ), Dongping TU(
), Dongping TU( )
)
Received:2024-04-14
Accepted:2024-06-04
Online:2025-10-15
Published:2025-10-15
Contact:
Liuping WANG,Dongping TU
陈秋洁( ), 黄玉妹, 冼金燕, 李凯童, 黄杰兰, 石小秀, 王柳萍(
), 黄玉妹, 冼金燕, 李凯童, 黄杰兰, 石小秀, 王柳萍( ), 涂冬萍(
), 涂冬萍( )
)
通讯作者:
王柳萍,涂冬萍
作者简介:陈秋洁 E-mail:2358518871@qq.com
基金资助:CLC Number:
Qiujie CHEN, Yumei HUANG, Jinyan XIAN, Kaitong LI, Jielan HUANG, Xiaoxiu SHI, Liuping WANG, Dongping TU. Bioinformatics and Tissue-specific Expression Analysis of YUCCA Gene Family in Huperzia serrata[J]. Journal of Agricultural Science and Technology, 2025, 27(10): 72-83.
陈秋洁, 黄玉妹, 冼金燕, 李凯童, 黄杰兰, 石小秀, 王柳萍, 涂冬萍. 蛇足石杉YUCCA基因家族的生物信息学及其组织特异表达分析[J]. 中国农业科技导报, 2025, 27(10): 72-83.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2024.0297
网站平台 Website platform | 网址 Website | 功能 Function |
|---|---|---|
| BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | 序列比对筛选基因 Screening genes by sequence alignment |
| ORF Finder | https://www.ncbi.nlm.nih.gov/orffinder | 获得潜在的蛋白质编码片段 Obtain potential protein-coding fragments |
| SMART | http://smart.embl-heidelberg.de/ | 结构域预测 Domain prediction |
| InterPro | https://www.ebi.ac.uk/interpro/search/sequence/ | 结构域预测 Domain prediction |
| Batch CD-Search | https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | 结构域预测 Domain prediction |
| ExPAsy | https://web.expasy.org/protparam/ | 理化性质分析 Analysis of physicochemical property |
| Plant-mPLoc | http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ | 亚细胞定位预测 Subcellular localization prediction |
| DeepTMHMM | https://dtu.biolib.com/DeepTMHMM | 跨膜预测 Transmembrane prediction |
| SignalP-6.0 | https://services.healthtech.dtu.dk/services/SignalP-6.0/ | 信号肽预测 Signal peptide prediction |
| Prabi | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html | 二级结构预测 Secondary structure prediction |
| Phyre2 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index | 三级结构预测 Tertiary structure prediction |
| MEME Suite 5.5.5 | https://meme-suite.org/meme/tools/meme | 保守基序预测 Conserved motif prediction |
| Plantcare | https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ | 顺式作用元件预测 Cis-acting element prediction |
Table 1 Main tools of bioinformatics analysis
网站平台 Website platform | 网址 Website | 功能 Function |
|---|---|---|
| BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | 序列比对筛选基因 Screening genes by sequence alignment |
| ORF Finder | https://www.ncbi.nlm.nih.gov/orffinder | 获得潜在的蛋白质编码片段 Obtain potential protein-coding fragments |
| SMART | http://smart.embl-heidelberg.de/ | 结构域预测 Domain prediction |
| InterPro | https://www.ebi.ac.uk/interpro/search/sequence/ | 结构域预测 Domain prediction |
| Batch CD-Search | https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | 结构域预测 Domain prediction |
| ExPAsy | https://web.expasy.org/protparam/ | 理化性质分析 Analysis of physicochemical property |
| Plant-mPLoc | http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ | 亚细胞定位预测 Subcellular localization prediction |
| DeepTMHMM | https://dtu.biolib.com/DeepTMHMM | 跨膜预测 Transmembrane prediction |
| SignalP-6.0 | https://services.healthtech.dtu.dk/services/SignalP-6.0/ | 信号肽预测 Signal peptide prediction |
| Prabi | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html | 二级结构预测 Secondary structure prediction |
| Phyre2 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index | 三级结构预测 Tertiary structure prediction |
| MEME Suite 5.5.5 | https://meme-suite.org/meme/tools/meme | 保守基序预测 Conserved motif prediction |
| Plantcare | https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ | 顺式作用元件预测 Cis-acting element prediction |
蛋白 Protein | 氨基酸数量Number of amino acids/aa | 分子量 Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂溶指数Aliphatic index | 平均亲水性指数 GRAVY |
|---|---|---|---|---|---|---|
| HsYUCCA1 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA2 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA3 | 167 | 17 161.46 | 5.30 | 41.09 | 86.59 | 0.166 |
| HsYUCCA4 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA5 | 410 | 44 864.99 | 8.54 | 45.29 | 77.12 | -0.260 |
| HsYUCCA6 | 410 | 44 864.99 | 8.54 | 45.29 | 77.12 | -0.260 |
| HsYUCCA7 | 150 | 16 105.53 | 8.83 | 54.93 | 85.93 | -0.198 |
| HsYUCCA8 | 247 | 27 058.52 | 4.58 | 40.04 | 92.83 | -0.039 |
| HsYUCCA9 | 244 | 28 655.83 | 7.56 | 28.72 | 88.65 | -0.505 |
| HsYUCCA10 | 212 | 24 521.68 | 5.60 | 42.25 | 63.02 | -0.804 |
| HsYUCCA11 | 407 | 46 074.16 | 9.46 | 27.67 | 77.57 | -0.354 |
| HsYUCCA12 | 197 | 22 763.87 | 9.63 | 23.84 | 105.33 | 0.226 |
| HsYUCCA13 | 150 | 16 105.53 | 8.83 | 54.93 | 85.93 | -0.198 |
| HsYUCCA14 | 354 | 28 602.97 | 4.62 | 42.33 | 97.57 | -0.019 |
| HsYUCCA15 | 161 | 17 571.91 | 4.36 | 48.24 | 85.59 | -0.081 |
| HsYUCCA16 | 330 | 37 721.64 | 9.01 | 44.11 | 80.12 | -0.488 |
| HsYUCCA17 | 544 | 59 058.24 | 4.82 | 40.04 | 98.47 | 0.011 |
| HsYUCCA18 | 150 | 17 856.82 | 9.22 | 38.15 | 104.73 | -0.191 |
Table 2 Physicochemical characteristics ofHsYUCCA proteins
蛋白 Protein | 氨基酸数量Number of amino acids/aa | 分子量 Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂溶指数Aliphatic index | 平均亲水性指数 GRAVY |
|---|---|---|---|---|---|---|
| HsYUCCA1 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA2 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA3 | 167 | 17 161.46 | 5.30 | 41.09 | 86.59 | 0.166 |
| HsYUCCA4 | 207 | 22 567.66 | 5.97 | 45.77 | 83.04 | -0.338 |
| HsYUCCA5 | 410 | 44 864.99 | 8.54 | 45.29 | 77.12 | -0.260 |
| HsYUCCA6 | 410 | 44 864.99 | 8.54 | 45.29 | 77.12 | -0.260 |
| HsYUCCA7 | 150 | 16 105.53 | 8.83 | 54.93 | 85.93 | -0.198 |
| HsYUCCA8 | 247 | 27 058.52 | 4.58 | 40.04 | 92.83 | -0.039 |
| HsYUCCA9 | 244 | 28 655.83 | 7.56 | 28.72 | 88.65 | -0.505 |
| HsYUCCA10 | 212 | 24 521.68 | 5.60 | 42.25 | 63.02 | -0.804 |
| HsYUCCA11 | 407 | 46 074.16 | 9.46 | 27.67 | 77.57 | -0.354 |
| HsYUCCA12 | 197 | 22 763.87 | 9.63 | 23.84 | 105.33 | 0.226 |
| HsYUCCA13 | 150 | 16 105.53 | 8.83 | 54.93 | 85.93 | -0.198 |
| HsYUCCA14 | 354 | 28 602.97 | 4.62 | 42.33 | 97.57 | -0.019 |
| HsYUCCA15 | 161 | 17 571.91 | 4.36 | 48.24 | 85.59 | -0.081 |
| HsYUCCA16 | 330 | 37 721.64 | 9.01 | 44.11 | 80.12 | -0.488 |
| HsYUCCA17 | 544 | 59 058.24 | 4.82 | 40.04 | 98.47 | 0.011 |
| HsYUCCA18 | 150 | 17 856.82 | 9.22 | 38.15 | 104.73 | -0.191 |
蛋白 Protein | 亚细胞定位 Subcellular localization | 信号肽 Signal peptide | 跨膜Transmembrane |
|---|---|---|---|
| HsYUCCA1 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA2 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA3 | 叶绿体 Chloroplast | 无No | 1 |
| HsYUCCA4 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA5 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA6 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA7 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA8 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA9 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA10 | 细胞质,过氧化物酶体 Cytoplasm, peroxisome | 无No | 0 |
| HsYUCCA11 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA12 | 细胞膜 Cell membrane | 无No | 2 |
| HsYUCCA13 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA14 | 叶绿体,细胞核 Chloroplast, nucleus | 无No | 0 |
| HsYUCCA15 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA16 | 叶绿体 Chloroplast | 无No | 0 |
| HsYUCCA17 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA18 | 细胞核 Nucleus | 无No | 0 |
Table 3 Subcellular localization and signal peptide prediction of HsYUCCAs proteins
蛋白 Protein | 亚细胞定位 Subcellular localization | 信号肽 Signal peptide | 跨膜Transmembrane |
|---|---|---|---|
| HsYUCCA1 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA2 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA3 | 叶绿体 Chloroplast | 无No | 1 |
| HsYUCCA4 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA5 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA6 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA7 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA8 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA9 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA10 | 细胞质,过氧化物酶体 Cytoplasm, peroxisome | 无No | 0 |
| HsYUCCA11 | 叶绿体,细胞质 Chloroplast, cytoplasm | 无No | 0 |
| HsYUCCA12 | 细胞膜 Cell membrane | 无No | 2 |
| HsYUCCA13 | 线粒体 Mitochondrion | 无No | 1 |
| HsYUCCA14 | 叶绿体,细胞核 Chloroplast, nucleus | 无No | 0 |
| HsYUCCA15 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA16 | 叶绿体 Chloroplast | 无No | 0 |
| HsYUCCA17 | 细胞核 Nucleus | 无No | 0 |
| HsYUCCA18 | 细胞核 Nucleus | 无No | 0 |
蛋白 Protein | 建模氨基酸残基数量(比例) Modeling amino acids residues number (proportion/%) | 模型 Model | 序列同源性 Sequence homology/% | 类型 Type |
|---|---|---|---|---|
| HsYUCCA1 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA2 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA3 | 99(59) | c6l9sA | 99.8 | 金属结合蛋白 Metal binding protein |
| HsYUCCA4 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA5 | 284(69) | c8ewuB | 100.0 | 生物合成蛋白,氧化还原酶Biosynthetic protein, oxidoreductase |
| HsYUCCA6 | 284(69) | c8ewuB | 100.0 | 生物合成蛋白,氧化还原酶Biosynthetic protein, oxidoreductase |
| HsYUCCA7 | 144(96) | c4uzyA | 99.7 | 运动蛋白 Motor protein |
| HsYUCCA8 | 92(37) | c3q5xA | 4.2 | 细胞周期 Cell cycle |
| HsYUCCA9 | 237(97) | c4bzbB | 100.0 | 水解酶 Hydrolase |
| HsYUCCA10 | 199(94) | c5j7xA | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA11 | 330(81) | c5mq6A | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA12 | 69(35) | c6ttjC | 8.1 | 水解酶 Hydrolase |
| HsYUCCA13 | 144(96) | c4uzyA | 99.7 | 运动蛋白 Motor protein |
| HsYUCCA14 | 260(73) | c6wc3A | 96.0 | 转运蛋白 Transport protein |
| HsYUCCA15 | 128(79) | c5h11A | 91.6 | 胞吐 Exocytosis |
| HsYUCCA16 | 317(96) | c3uclA | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA17 | 494(90) | c5h11A | 98.1 | 胞吐 Exocytosis |
| HsYUCCA18 | 117(78) | c4bzbB | 100.0 | 水解酶 Hydrolase |
Table 4 Tertiary structure prediction of HsYUCCAs proteins
蛋白 Protein | 建模氨基酸残基数量(比例) Modeling amino acids residues number (proportion/%) | 模型 Model | 序列同源性 Sequence homology/% | 类型 Type |
|---|---|---|---|---|
| HsYUCCA1 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA2 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA3 | 99(59) | c6l9sA | 99.8 | 金属结合蛋白 Metal binding protein |
| HsYUCCA4 | 139(67) | c4eqfA | 99.7 | 转运蛋白 Transport protein |
| HsYUCCA5 | 284(69) | c8ewuB | 100.0 | 生物合成蛋白,氧化还原酶Biosynthetic protein, oxidoreductase |
| HsYUCCA6 | 284(69) | c8ewuB | 100.0 | 生物合成蛋白,氧化还原酶Biosynthetic protein, oxidoreductase |
| HsYUCCA7 | 144(96) | c4uzyA | 99.7 | 运动蛋白 Motor protein |
| HsYUCCA8 | 92(37) | c3q5xA | 4.2 | 细胞周期 Cell cycle |
| HsYUCCA9 | 237(97) | c4bzbB | 100.0 | 水解酶 Hydrolase |
| HsYUCCA10 | 199(94) | c5j7xA | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA11 | 330(81) | c5mq6A | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA12 | 69(35) | c6ttjC | 8.1 | 水解酶 Hydrolase |
| HsYUCCA13 | 144(96) | c4uzyA | 99.7 | 运动蛋白 Motor protein |
| HsYUCCA14 | 260(73) | c6wc3A | 96.0 | 转运蛋白 Transport protein |
| HsYUCCA15 | 128(79) | c5h11A | 91.6 | 胞吐 Exocytosis |
| HsYUCCA16 | 317(96) | c3uclA | 100.0 | 氧化还原酶 Oxidoreductase |
| HsYUCCA17 | 494(90) | c5h11A | 98.1 | 胞吐 Exocytosis |
| HsYUCCA18 | 117(78) | c4bzbB | 100.0 | 水解酶 Hydrolase |
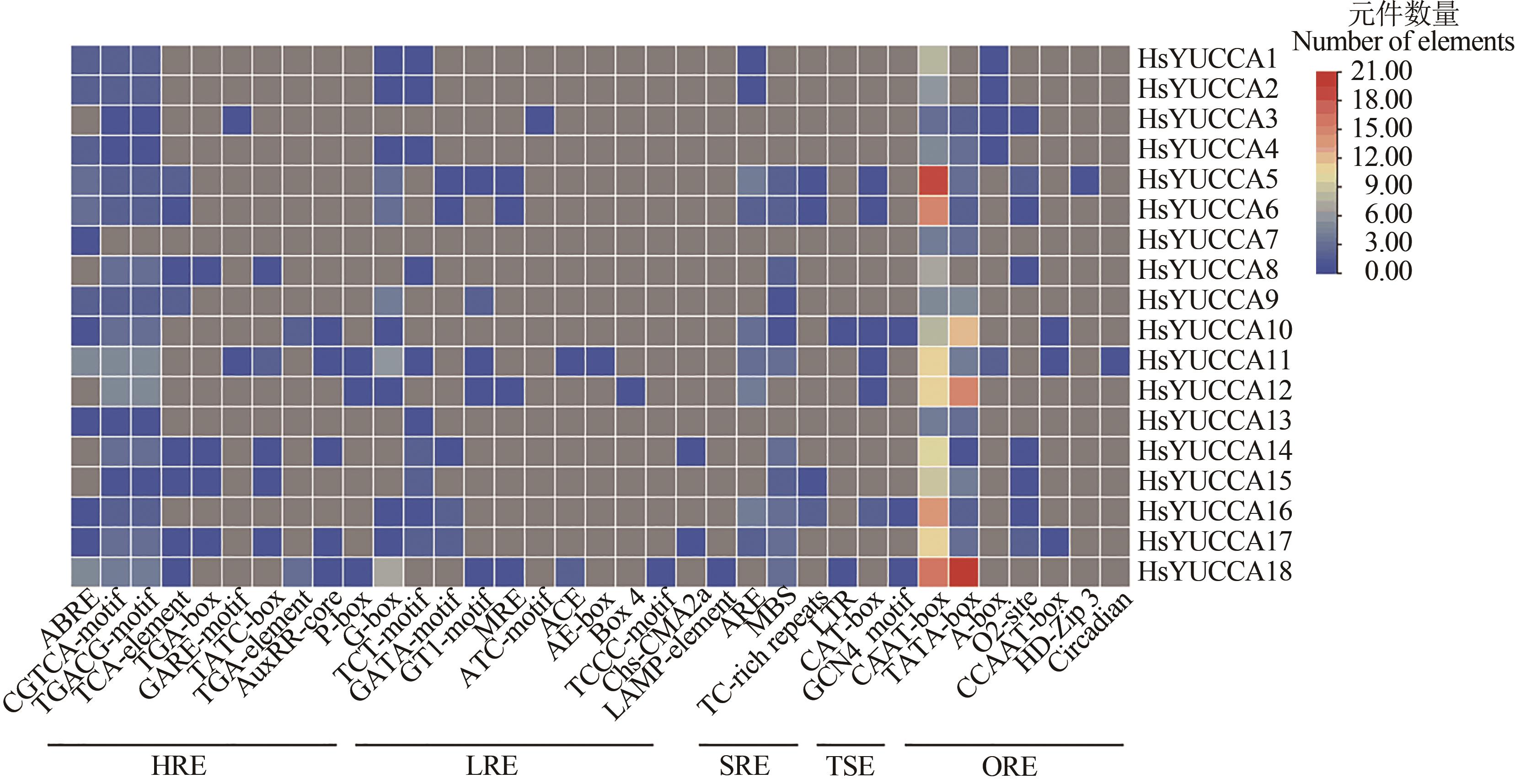
Fig. 5 Cis-acting elements heat map of HsYUCCA genefamilyNote:HRE—Hormone responsive elements; LRE—Light responsive elements; SRE—Stress responsive elements; TSE—Tissue specific elements; ORE—Other responsive elements.
| [1] | SAKATA T, YAGIHASHI N, HIGASHITANI A. Tissue-specific auxin signaling in response to temperature fluctuation [J]. Plant Signal. Behav., 2010, 5(11): 1510-1512. |
| [2] | WON C, SHEN X, MASHIGUCHI K,et al.. Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis [J]. Proc. Natl. Acad. Sci. USA, 2011, 108(45): 18518-18523. |
| [3] | MASHIGUCHI K, TANAKA K, SAKAI T, et al.. The main auxin biosynthesis pathway in Arabidopsis [J]. Proc. Natl. Acad.Sci. USA, 2011, 108(45): 18512-18517. |
| [4] | DI D W, WU L, ZHANG L, et al.. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin [J/OL]. Sci. Rep., 2016, 6: 36866 [2024-03-10]. . |
| [5] | ZHAO Y, CHRISTENSEN S K, FANKHAUSER C, et al.. A role for flavin monooxygenase-like enzymes in auxin biosynthesis [J]. Viruses, 2001, 291(5502): 306-309. |
| [6] | ROSS J J, TIVENDALE N D, REID J B, et al.. Reassessing the role of YUCCAs in auxin biosynthesis [J]. Plant Signal. Behav.,2011, 6(3): 437-439. |
| [7] | OVERVOORDE P, FUKAKI H, BEECKMAN T. Auxin control of root development [J/OL]. Cold Spring Harb. Perspect. Biol.,2010, 2(6): a001537 [2024-03-10]. . |
| [8] | CAO X, YANG H, SHANG C, et al.. The roles of auxin biosynthesis YUCCA gene family in plants [J/OL]. Int. J. Mol.Sci., 2019, 20(24): E6343 [2024-03-10]. . |
| [9] | GÉLINAS-MARION A, ELÉOUËT M P, COOK S D, et al..Plant development in the garden pea as revealed by mutations in the Crd/PsYUC1 gene [J/OL].Genes, 2023, 14(12): 2115 [2024-03-10]. . |
| [10] | ZHANG Y H, YIN H, ZHAO X M, et al.. The promoting effects of alginate oligosaccharides on root development in Oryza sativa L.mediated by auxin signaling [J]. Carbohydr. Polym.,2014, 113: 446-454. |
| [11] | 钟海英, 罗义, 龚钰华, 等. RNAi法分析香菇YUCCA8基因的功能[J]. 食用菌学报, 2021, 28(3): 11-18. |
| ZHONG H Y, LUO Y, GONG Y H, et al.. Functional analysis of Indole-3-pyruvate monooxygenase gene YUCCA8 involved in heat resistance of Lentinula edodes by RNAi [J]. Acta Edulis Fungi, 2021, 28(3): 11-18. | |
| [12] | 王晶.菘蓝IiYUCCA6基因的克隆及其功能研究[D]. 西安: 陕西师范大学, 2018. |
| WANG J. Cloning and functional analysis of IiYUCCA6 gene in Isatis indigotica [D]. Xi'an: Shaanxi Normal University, 2018. | |
| [13] | 蒋日红, 张宪春. 广西石松科植物新记录及更正[J]. 广西林业科学, 2022, 51(3): 452-454. |
| JIANG R H, ZHANG X C.New records and corrections of Lycopodiaceae from Guangxi [J]. Guangxi For. Sci., 2022, 51(3): 452-454. | |
| [14] | 李籽杉, 安周捷, 王婧, 等. 蛇足石杉主要活性成分及其生物合成研究进展[J]. 中草药, 2022, 53(11): 3505-3517. |
| LI Z S, AN Z J, WANG J, et al.. Active compounds and their biosynthesis of Huperzia serrate [J]. Chin.Tradit.Herb.Drugs,2022, 53(11): 3505-3517. | |
| [15] | 黄玉妹, 谢采连, 周玲梅, 等. 千层塔基原及药效文献研究[J]. 亚太传统医药, 2023, 19(3): 183-189. |
| HUANG Y M, XIE C L, ZHOU L M, et al.. Literature research on the original and efficacy of Huperzia serrate (Thunb.) Trev. [J]. Asia-Pacific Tradit. Med., 2023, 19(3): 183-189. | |
| [16] | 王飞. 石杉碱甲片治疗阿尔茨海默症的疗效和安全性[J]. 中国老年保健医学, 2017, 15(6): 67-68. |
| [17] | 张健, 李全, 周妍妍. 中药调控Nrf2通路防治阿尔茨海默病的研究进展[J]. 中国实验方剂学杂志, 2024, 30(9): 219-227. |
| ZHANG J, LI Q, ZHOU Y Y. Research progress of traditional chinese medicine regulating Nrf2 pathway in the prevention and treatment of Alzheimer's disease [J]. Chin. J. Exp. Tradit. Med. Form., 2024, 30(9): 219-227. | |
| [18] | BREIJYEH Z, KARAMAN R. Comprehensive review on Alzheimer's disease:causes and treatment [J/OL].Molecules (Basel Switz.), 2020, 25(24): E5789 [2024-03-10]. . |
| [19] | 黄玉妹, 滕建北, 涂冬萍, 等. 蛇足石杉COBRA基因家族的分子生物信息学及表达分析[J]. 广西植物, 2024, 44(6):1105-1117. |
| HUANG Y M, TENG J B, TU D P, et al.. Molecular bioinformatics and expression analysis of the COBRA gene family in Huperzia serrata [J]. Guihaia, 2024, 44(6): 1105-1117. | |
| [20] | 李海波, 史济东, 张恺, 等.基于全长转录组的蛇足石杉ARF基因家族鉴定分析[J].广西植物, 2024, 44(6): 1091-1104. |
| LI H B, SHI J D, ZHANG K, et al.. Identification and analysis of the ARF gene family based on full-length transcriptome in Huperzia serrata [J]. Guihaia, 2024, 44(6): 1091-1104. | |
| [21] | YANG Y L, XU T, WANG H G,et al.. Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat [J]. Mol. Biol. Rep., 2021, 48(2): 1269-1279. |
| [22] | 刘洪.草莓YUCCA基因家族的分离与基因功能研究[D]. 南京:南京农业大学, 2012. |
| LIU H. Molecular characterization and functional analyisis of the YUCCA flavin monooxygenase genes from strawberry (Fragaria L.) [D]. Nanjing: Nanjing Agricultural University, 2012. | |
| [23] | 张倩倩, 田守蔚, 张洁, 等. 西瓜YUCCA基因家族鉴定及在果实成熟过程中的表达分析[J].中国蔬菜,2019(3):21-29. |
| ZHANG Q Q, TIAN S W, ZHANG J, et al.. Identification of YUCCA gene family and expression analysis during watermelon fruit ripening process [J]. China Veg., 2019(3): 21-29. | |
| [24] | 张燕萍, 童妍, 胡美娟, 等. 小兰屿蝴蝶兰YUCCA基因家族鉴定及在花朵中的表达分析[J]. 分子植物育种, 2024,22(13): 4177-4184. |
| ZHANG Y P, TONG Y, HU M J, et al.. Identification of YUCCA gene family and analysis of its expression in flowers of Phalaenopsis equestris [J]. Mol. Plant Breeding, 2024, 22(13):4177-4184. | |
| [25] | 褚晶, 田晓芹, 陈世华, 等.藜麦黄素单加氧酶基因(CqYUCCA)家族的全基因组鉴定与分析[J]. 烟台大学学报(自然科学与工程版), 2023, 36(1): 20-27. |
| CHU J, TIAN X Q, CHEN S H, et al.. Genome-wide identification and analysis of flavin monooxygenase gene (CqYUCCA) family in quinoa (Chenopodium quinoa Willd.) [J].J. Yantai Univ. (Nat. Sci. Eng.), 2023, 36(1): 20-27. | |
| [26] | 王仁汉, 宋志美, 屈旭, 等. 普通烟草YUCCA基因家族的生物信息学分析[J]. 江苏农业科学, 2021, 49(3): 61-65. |
| [27] | 莫福磊, 束艺, 陈秀玲, 等. 基于全基因组的番茄YUCCA基因家族生物信息学分析[J]. 分子植物育种, 2020, 18(10):3159-3163. |
| MO F L, SHU Y, CHEN X L, et al.. Bioinformatics analysis of tomato YUCCA gene family based on whole genome [J]. Mol.Plant Breed., 2020, 18(10): 3159-3163. | |
| [28] | 袁美同, 李绍信, 纪丕钰, 等. 梨YUCCA基因家族的鉴定与生物信息学分析[J]. 分子植物育种, 2021, 19(19): 6328-6337. |
| YUAN M T, LI S X, JI P Y, et al.. Identification and bioinformatics analysis of YUCCA gene family in Pyrus [J]. Mol.Plant Breed., 2021, 19(19): 6328-6337. | |
| [29] | 闫学敏, 吴英华, 史艳, 等. 胡萝卜YUCCA基因家族鉴定及生物信息学分析[J]. 江苏农业科学, 2021, 49(23): 52-57. |
| [30] | SONG C H, ZHANG D, ZHENG L W, et al.. Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica [J/OL]. Sci.Rep., 2020, 10:10866 [2024-03-10]. . |
| [31] | 綦洋, 王柬钧, 桑园园, 等. 大白菜YUCCA基因家族的鉴定与生物信息学分析[J]. 江苏农业科学, 2019, 47(3): 49-54. |
| [32] | 杨昌华, 胡文胜, 李树华, 等. 石杉碱甲新型衍生物的合成及其对神经细胞活性的影响[J]. 湖南师范大学学报(医学版), 2023, 20(3): 155-157. |
| YANG C H, HU W S, LI S H, et al.. Synthesis of novel (-)-huperzine A derivatives and the effects on nerve cell viability [J]. J. Hunan Norm. Univ. (Med. Sci.), 2023, 20(3): 155-157. | |
| [33] | 曹朵. 千层塔抗阿尔茨海默氏病的物质基础及其作用机制研究[D]. 西安: 西北大学, 2022. |
| CAO D. Study on the effective constituents and mechanism of Huperzia serrata against alzheimer's disease [D]. Xi'an: Northwest University, 2022. | |
| [34] | 陈思思, 张梦华, 王锦秀, 等. 药用植物千层塔的基原物种及研究进展[J]. 广西植物, 2021, 41(11): 1794-1809. |
| CHEN S S, ZHANG M H, WANG J X, et al.. Original plant and research progress of the medicinal plant Huperzia javanica [J]. Guihaia, 2021, 41(11): 1794-1809. | |
| [35] | KIM J I, BAEK D, PARK H C, et al.. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit [J]. Mol.Plant, 2013, 6(2): 337-349. |
| [36] | 徐玲玲, 陈习, 易立, 等. 千层塔形态与石杉碱甲含量的关系研究[J]. 山东农业科学, 2013, 45(9): 36-38. |
| XU L L, CHEN X, YI L, et al.. Study on relationship between morphology and huperzine A content of Huperzia serrate [J].Shandong Agric. Sci., 2013, 45(9): 36-38. | |
| [37] | YAMAMOTO Y, KAMIYA N, MORINAKA Y, et al.. Auxin biosynthesis by the YUCCA genes in rice [J]. Plant Physiol.,2007, 143(3): 1362-1371. |
| [38] | HENTRICH M, BÖTTCHER C, DÜCHTING P, et al.. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression [J]. Plant J., 2013, 74(4): 626-637. |
| [1] | Zhien LIU, Yong HE, Zhicheng WANG, Xiaokang ZHAN, Tingbao WANG, Yaowei LIU, Zhihong TIAN. Identification and Bioinformatics Analysis of Growth Regulating Factor GRF Gene Family in Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 18-27. |
| [2] | Zhining YANG, Xuke LU, Yapeng FAN, Yuping SUN, Xin YU, Liang WANG, Yongjian GAO, Gulijiayinashen Habuli, Gulishaxi Nasiyi, Wumuer Abudu, Hui HUANG, Menghao ZHANG, Lidong WANG, Xiao CHEN, Lei XIAO, Xinrui ZHANG, Shuai WANG, Xiugui CHEN, Junjuan WANG, Lixue GUO, Wenwei GAO, Wuwei YE. Response Mechanism of Cotton GhDMT7 Gene to 5-azacytidine [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 28-35. |
| [3] | Jinxian LIU, Lijuan WANG, Jie LIU, Xianyu FU, Guangheng WU. Identification and Expression Analysis of Calmodulin-binding Transcription Activator (CAMTA) Family Genes in Tea Plants [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 71-82. |
| [4] | Pan ZHANG, Haichao SUN, Wenheng DONG, Yongjiang LI, Yingying ZHANG, Lili SHI, Jun ZHAO, Daowen LU. Genome-wide Identification and Expression Analysis of Maize JRL Gene Family [J]. Journal of Agricultural Science and Technology, 2025, 27(10): 32-41. |
| [5] | Xianguo LI, Qi DAI, Zepeng WANG, Zhaolong CHEN, Huizhuan YAN, Ning LI. Identification and Phylogenetic Analysis of Tomato CCCH-like Zinc Finger Protein Family [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 80-95. |
| [6] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [7] | Fulin ZHANG, Rui XI, Yuxiang LIU, Zhaolong CHEN, Qinghui YU, Ning LI. Genome-wide Identification and Expression Analysis of Tomato BURP Structural Domain Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 51-62. |
| [8] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [9] | Mingdi CHEN, Guihua HU, Haiwen ZHANG, Wangtian WANG. Bioinformatics and Expression Pattern Analysis of Rice RR Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 20-29. |
| [10] | Zhanqing WU, Wei CHEN, Zhan ZHAO, Hailiang XU, Haoyuan LI, Xingxing PENG, Dongxu CHEN, Mingyue ZHANG. Genome-wide Identification and Bioinformatics Analysis of GRAS Gene Family in Maize [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 15-25. |
| [11] | Bo LIU, Wangtian WANG, Li MA, Junyan WU, Yuanyuan PU, Lijun LIU, Yan FANG, Wancang SUN, Yan ZHANG, Ruimin LIU, Xiucun ZENG. Identification and Characterization of IPT Gene Family in Brassica rapa L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 56-66. |
| [12] | Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN, Gengqing HUANG. Bioinformatics and Expression Pattern Analysis of JAZ and MYC2-like Gene Family in Lavandula angustifolia [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 79-90. |
| [13] | Zhenwei ZHANG, Xiangshu DONG, Jing YANG, Xuejun LI, Meijun QI, Kuaile JIANG, Yonglin YANG, Butian WANG, Xuedong SHI, Junchao QIU, Zhihua CHEN, Yu GE. Genome-wide Identification and Expression Analysis of Key Chlorophyll Synthesis Related Gene CaPOR in Coffea arabica [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 83-97. |
| [14] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [15] | Jinfeng ZHAO, Aili YU, Yanfang LI, Yanwei DU, Gaohong WANG, Zhenhua WANG. Response Characteristics of SiCBL3 to Abiotic Stresses in Foxtail Millet [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 68-75. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号