




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (3): 15-25.DOI: 10.13304/j.nykjdb.2023.0551
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Zhanqing WU1( ), Wei CHEN1(
), Wei CHEN1( ), Zhan ZHAO2, Hailiang XU1, Haoyuan LI1, Xingxing PENG1, Dongxu CHEN1, Mingyue ZHANG1
), Zhan ZHAO2, Hailiang XU1, Haoyuan LI1, Xingxing PENG1, Dongxu CHEN1, Mingyue ZHANG1
Received:2023-07-17
Accepted:2023-09-08
Online:2024-03-15
Published:2024-03-07
Contact:
Wei CHEN
吴占清1( ), 陈威1(
), 陈威1( ), 赵展2, 许海良1, 李豪远1, 彭星星1, 陈东旭1, 张明月1
), 赵展2, 许海良1, 李豪远1, 彭星星1, 陈东旭1, 张明月1
通讯作者:
陈威
作者简介:吴占清 E-mail:13383782062@163.com;
基金资助:CLC Number:
Zhanqing WU, Wei CHEN, Zhan ZHAO, Hailiang XU, Haoyuan LI, Xingxing PENG, Dongxu CHEN, Mingyue ZHANG. Genome-wide Identification and Bioinformatics Analysis of GRAS Gene Family in Maize[J]. Journal of Agricultural Science and Technology, 2024, 26(3): 15-25.
吴占清, 陈威, 赵展, 许海良, 李豪远, 彭星星, 陈东旭, 张明月. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析[J]. 中国农业科技导报, 2024, 26(3): 15-25.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2023.0551
基因名称 Gene name | 基因ID Gene ID | 氨基酸长度Amino acids length/aa | 分子量Molecular weight/Da | 等电点PI | 带负电荷残基总数 Total number of negatively charged residues | 带正电荷残基总数 Total number of positively charged residues | 不稳定 指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 平均 疏水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|---|---|
| ZmGRAS1 | Zm00001eb216610 | 625 | 65 742.60 | 5.18 | 70 | 50 | 46.59 | 75.66 | -0.204 |
| ZmGRAS2 | Zm00001eb107910 | 554 | 60 835.57 | 5.84 | 68 | 52 | 53.95 | 83.83 | -0.302 |
| ZmGRAS3 | Zm00001eb325750 | 558 | 60 991.58 | 5.71 | 69 | 52 | 55.68 | 82.17 | -0.301 |
| ZmGRAS4 | Zm00001eb106490 | 570 | 64 511.14 | 5.99 | 73 | 64 | 46.97 | 83.72 | -0.475 |
| ZmGRAS5 | Zm00001eb027720 | 542 | 59 914.41 | 5.76 | 58 | 43 | 48.73 | 83.86 | -0.245 |
| ZmGRAS6 | Zm00001eb170110 | 775 | 82 511.74 | 6.19 | 73 | 62 | 63.30 | 74.27 | -0.245 |
| ZmGRAS7 | Zm00001eb195650 | 668 | 71 162.84 | 6.15 | 66 | 53 | 57.35 | 85.21 | -0.197 |
| ZmGRAS8 | Zm00001eb288400 | 561 | 62 826.62 | 4.92 | 79 | 54 | 46.74 | 81.23 | -0.340 |
| ZmGRAS9 | Zm00001eb141310 | 447 | 48 471.03 | 6.36 | 55 | 50 | 54.57 | 89.44 | -0.145 |
| ZmGRAS10 | Zm00001eb093670 | 678 | 72 083.67 | 6.18 | 67 | 55 | 56.48 | 87.23 | -0.168 |
| ZmGRAS11 | Zm00001eb363140 | 449 | 48 889.56 | 6.26 | 56 | 50 | 56.39 | 89.49 | -0.133 |
| ZmGRAS12 | Zm00001eb148290 | 809 | 89 139.89 | 6.06 | 101 | 88 | 44.93 | 77.00 | -0.344 |
| ZmGRAS13 | Zm00001eb378590 | 616 | 65 472.30 | 6.30 | 70 | 63 | 45.04 | 71.62 | -0.240 |
| ZmGRAS14 | Zm00001eb136350 | 710 | 78 391.49 | 5.30 | 92 | 73 | 53.77 | 79.77 | -0.339 |
| ZmGRAS15 | Zm00001eb324600 | 452 | 48 411.17 | 6.05 | 55 | 49 | 58.80 | 96.26 | -0.044 |
| ZmGRAS16 | Zm00001eb164530 | 765 | 84 654.31 | 5.30 | 103 | 78 | 49.74 | 73.02 | -0.392 |
| ZmGRAS17 | Zm00001eb055700 | 551 | 59 842.10 | 5.20 | 63 | 47 | 56.67 | 86.03 | -0.091 |
| ZmGRAS18 | Zm00001eb215870 | 569 | 61 233.71 | 5.08 | 63 | 44 | 57.67 | 85.59 | -0.056 |
| ZmGRAS19 | Zm00001eb344400 | 508 | 56 088.05 | 5.91 | 55 | 43 | 35.75 | 90.08 | -0.105 |
| ZmGRAS20 | Zm00001eb164480 | 642 | 71 250.62 | 6.67 | 84 | 80 | 52.59 | 79.63 | -0.405 |
| ZmGRAS21 | Zm00001eb164490 | 666 | 73 486.54 | 5.51 | 93 | 76 | 51.16 | 74.25 | -0.425 |
| ZmGRAS22 | Zm00001eb326020 | 592 | 63 904.56 | 5.67 | 64 | 50 | 58.88 | 68.06 | -0.370 |
| ZmGRAS23 | Zm00001eb108250 | 456 | 49 134.09 | 5.32 | 50 | 35 | 40.45 | 90.99 | 0.057 |
| ZmGRAS24 | Zm00001eb011560 | 564 | 59 835.16 | 5.12 | 65 | 44 | 43.97 | 75.30 | -0.152 |
| ZmGRAS25 | Zm00001eb326210 | 456 | 49 478.48 | 5.38 | 51 | 36 | 39.36 | 89.06 | 0.011 |
| ZmGRAS26 | Zm00001eb108090 | 586 | 63 755.58 | 5.49 | 66 | 53 | 59.45 | 70.56 | -0.343 |
| ZmGRAS27 | Zm00001eb081760 | 607 | 66 678.45 | 6.04 | 77 | 67 | 47.34 | 80.97 | -0.287 |
| ZmGRAS28 | Zm00001eb020850 | 630 | 67 547.26 | 6.15 | 65 | 53 | 66.00 | 63.40 | -0.416 |
| ZmGRAS29 | Zm00001eb073130 | 623 | 65 570.98 | 6.15 | 60 | 54 | 52.23 | 87.26 | -0.116 |
| ZmGRAS30 | Zm00001eb187310 | 607 | 63 032.09 | 8.75 | 54 | 59 | 46.96 | 84.93 | -0.109 |
| ZmGRAS31 | Zm00001eb428430 | 623 | 65 303.67 | 6.34 | 58 | 54 | 48.36 | 84.11 | -0.137 |
| ZmGRAS32 | Zm00001eb164470 | 645 | 72 101.21 | 6.37 | 78 | 70 | 47.76 | 80.96 | -0.356 |
| ZmGRAS33 | Zm00001eb165340 | 639 | 72 513.56 | 6.09 | 85 | 75 | 45.77 | 81.61 | -0.384 |
| ZmGRAS34 | Zm00001eb236120 | 416 | 43 484.86 | 5.43 | 48 | 35 | 32.57 | 88.92 | -0.042 |
| ZmGRAS35 | Zm00001eb291030 | 497 | 52 002.72 | 5.80 | 51 | 38 | 46.41 | 82.39 | -0.007 |
| ZmGRAS36 | Zm00001eb291930 | 426 | 45 514.68 | 6.09 | 48 | 43 | 47.88 | 87.37 | -0.100 |
| ZmGRAS37 | Zm00001eb115950 | 599 | 67 026.72 | 6.12 | 76 | 68 | 50.16 | 74.89 | -0.375 |
| ZmGRAS38 | Zm00001eb074460 | 721 | 75 225.55 | 5.74 | 62 | 46 | 47.98 | 82.98 | -0.050 |
| ZmGRAS39 | Zm00001eb429450 | 718 | 75 350.35 | 5.72 | 64 | 47 | 50.61 | 81.82 | -0.081 |
| ZmGRAS40 | Zm00001eb186770 | 708 | 73 942.10 | 5.63 | 65 | 48 | 45.89 | 87.67 | 0.004 |
| ZmGRAS41 | Zm00001eb000470 | 481 | 51 254.82 | 6.10 | 48 | 37 | 41.79 | 95.88 | 0.099 |
| ZmGRAS42 | Zm00001eb196430 | 487 | 51 456.32 | 6.18 | 59 | 53 | 54.58 | 89.08 | -0.151 |
| ZmGRAS43 | Zm00001eb249840 | 706 | 73 961.86 | 5.47 | 66 | 44 | 45.87 | 85.55 | 0.001 |
| ZmGRAS44 | Zm00001eb202270 | 473 | 51 700.38 | 4.73 | 62 | 32 | 52.48 | 88.12 | 0.013 |
| ZmGRAS45 | Zm00001eb326550 | 306 | 33 520.48 | 6.05 | 30 | 25 | 33.94 | 97.32 | 0.086 |
| ZmGRAS46 | Zm00001eb139470 | 492 | 52 614.47 | 5.15 | 58 | 39 | 47.00 | 87.11 | -0.015 |
| ZmGRAS47 | Zm00001eb019640 | 526 | 54 677.28 | 6.72 | 53 | 51 | 51.38 | 76.24 | -0.189 |
| ZmGRAS48 | Zm00001eb405080 | 235 | 25 649.61 | 6.55 | 27 | 25 | 48.23 | 99.66 | 0.063 |
| ZmGRAS49 | Zm00001eb002990 | 313 | 33 288.60 | 9.66 | 25 | 36 | 54.58 | 86.77 | 0.034 |
Table 1 Physicochemical properties of proteins encoded by the GRAS gene family in maize
基因名称 Gene name | 基因ID Gene ID | 氨基酸长度Amino acids length/aa | 分子量Molecular weight/Da | 等电点PI | 带负电荷残基总数 Total number of negatively charged residues | 带正电荷残基总数 Total number of positively charged residues | 不稳定 指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 平均 疏水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|---|---|
| ZmGRAS1 | Zm00001eb216610 | 625 | 65 742.60 | 5.18 | 70 | 50 | 46.59 | 75.66 | -0.204 |
| ZmGRAS2 | Zm00001eb107910 | 554 | 60 835.57 | 5.84 | 68 | 52 | 53.95 | 83.83 | -0.302 |
| ZmGRAS3 | Zm00001eb325750 | 558 | 60 991.58 | 5.71 | 69 | 52 | 55.68 | 82.17 | -0.301 |
| ZmGRAS4 | Zm00001eb106490 | 570 | 64 511.14 | 5.99 | 73 | 64 | 46.97 | 83.72 | -0.475 |
| ZmGRAS5 | Zm00001eb027720 | 542 | 59 914.41 | 5.76 | 58 | 43 | 48.73 | 83.86 | -0.245 |
| ZmGRAS6 | Zm00001eb170110 | 775 | 82 511.74 | 6.19 | 73 | 62 | 63.30 | 74.27 | -0.245 |
| ZmGRAS7 | Zm00001eb195650 | 668 | 71 162.84 | 6.15 | 66 | 53 | 57.35 | 85.21 | -0.197 |
| ZmGRAS8 | Zm00001eb288400 | 561 | 62 826.62 | 4.92 | 79 | 54 | 46.74 | 81.23 | -0.340 |
| ZmGRAS9 | Zm00001eb141310 | 447 | 48 471.03 | 6.36 | 55 | 50 | 54.57 | 89.44 | -0.145 |
| ZmGRAS10 | Zm00001eb093670 | 678 | 72 083.67 | 6.18 | 67 | 55 | 56.48 | 87.23 | -0.168 |
| ZmGRAS11 | Zm00001eb363140 | 449 | 48 889.56 | 6.26 | 56 | 50 | 56.39 | 89.49 | -0.133 |
| ZmGRAS12 | Zm00001eb148290 | 809 | 89 139.89 | 6.06 | 101 | 88 | 44.93 | 77.00 | -0.344 |
| ZmGRAS13 | Zm00001eb378590 | 616 | 65 472.30 | 6.30 | 70 | 63 | 45.04 | 71.62 | -0.240 |
| ZmGRAS14 | Zm00001eb136350 | 710 | 78 391.49 | 5.30 | 92 | 73 | 53.77 | 79.77 | -0.339 |
| ZmGRAS15 | Zm00001eb324600 | 452 | 48 411.17 | 6.05 | 55 | 49 | 58.80 | 96.26 | -0.044 |
| ZmGRAS16 | Zm00001eb164530 | 765 | 84 654.31 | 5.30 | 103 | 78 | 49.74 | 73.02 | -0.392 |
| ZmGRAS17 | Zm00001eb055700 | 551 | 59 842.10 | 5.20 | 63 | 47 | 56.67 | 86.03 | -0.091 |
| ZmGRAS18 | Zm00001eb215870 | 569 | 61 233.71 | 5.08 | 63 | 44 | 57.67 | 85.59 | -0.056 |
| ZmGRAS19 | Zm00001eb344400 | 508 | 56 088.05 | 5.91 | 55 | 43 | 35.75 | 90.08 | -0.105 |
| ZmGRAS20 | Zm00001eb164480 | 642 | 71 250.62 | 6.67 | 84 | 80 | 52.59 | 79.63 | -0.405 |
| ZmGRAS21 | Zm00001eb164490 | 666 | 73 486.54 | 5.51 | 93 | 76 | 51.16 | 74.25 | -0.425 |
| ZmGRAS22 | Zm00001eb326020 | 592 | 63 904.56 | 5.67 | 64 | 50 | 58.88 | 68.06 | -0.370 |
| ZmGRAS23 | Zm00001eb108250 | 456 | 49 134.09 | 5.32 | 50 | 35 | 40.45 | 90.99 | 0.057 |
| ZmGRAS24 | Zm00001eb011560 | 564 | 59 835.16 | 5.12 | 65 | 44 | 43.97 | 75.30 | -0.152 |
| ZmGRAS25 | Zm00001eb326210 | 456 | 49 478.48 | 5.38 | 51 | 36 | 39.36 | 89.06 | 0.011 |
| ZmGRAS26 | Zm00001eb108090 | 586 | 63 755.58 | 5.49 | 66 | 53 | 59.45 | 70.56 | -0.343 |
| ZmGRAS27 | Zm00001eb081760 | 607 | 66 678.45 | 6.04 | 77 | 67 | 47.34 | 80.97 | -0.287 |
| ZmGRAS28 | Zm00001eb020850 | 630 | 67 547.26 | 6.15 | 65 | 53 | 66.00 | 63.40 | -0.416 |
| ZmGRAS29 | Zm00001eb073130 | 623 | 65 570.98 | 6.15 | 60 | 54 | 52.23 | 87.26 | -0.116 |
| ZmGRAS30 | Zm00001eb187310 | 607 | 63 032.09 | 8.75 | 54 | 59 | 46.96 | 84.93 | -0.109 |
| ZmGRAS31 | Zm00001eb428430 | 623 | 65 303.67 | 6.34 | 58 | 54 | 48.36 | 84.11 | -0.137 |
| ZmGRAS32 | Zm00001eb164470 | 645 | 72 101.21 | 6.37 | 78 | 70 | 47.76 | 80.96 | -0.356 |
| ZmGRAS33 | Zm00001eb165340 | 639 | 72 513.56 | 6.09 | 85 | 75 | 45.77 | 81.61 | -0.384 |
| ZmGRAS34 | Zm00001eb236120 | 416 | 43 484.86 | 5.43 | 48 | 35 | 32.57 | 88.92 | -0.042 |
| ZmGRAS35 | Zm00001eb291030 | 497 | 52 002.72 | 5.80 | 51 | 38 | 46.41 | 82.39 | -0.007 |
| ZmGRAS36 | Zm00001eb291930 | 426 | 45 514.68 | 6.09 | 48 | 43 | 47.88 | 87.37 | -0.100 |
| ZmGRAS37 | Zm00001eb115950 | 599 | 67 026.72 | 6.12 | 76 | 68 | 50.16 | 74.89 | -0.375 |
| ZmGRAS38 | Zm00001eb074460 | 721 | 75 225.55 | 5.74 | 62 | 46 | 47.98 | 82.98 | -0.050 |
| ZmGRAS39 | Zm00001eb429450 | 718 | 75 350.35 | 5.72 | 64 | 47 | 50.61 | 81.82 | -0.081 |
| ZmGRAS40 | Zm00001eb186770 | 708 | 73 942.10 | 5.63 | 65 | 48 | 45.89 | 87.67 | 0.004 |
| ZmGRAS41 | Zm00001eb000470 | 481 | 51 254.82 | 6.10 | 48 | 37 | 41.79 | 95.88 | 0.099 |
| ZmGRAS42 | Zm00001eb196430 | 487 | 51 456.32 | 6.18 | 59 | 53 | 54.58 | 89.08 | -0.151 |
| ZmGRAS43 | Zm00001eb249840 | 706 | 73 961.86 | 5.47 | 66 | 44 | 45.87 | 85.55 | 0.001 |
| ZmGRAS44 | Zm00001eb202270 | 473 | 51 700.38 | 4.73 | 62 | 32 | 52.48 | 88.12 | 0.013 |
| ZmGRAS45 | Zm00001eb326550 | 306 | 33 520.48 | 6.05 | 30 | 25 | 33.94 | 97.32 | 0.086 |
| ZmGRAS46 | Zm00001eb139470 | 492 | 52 614.47 | 5.15 | 58 | 39 | 47.00 | 87.11 | -0.015 |
| ZmGRAS47 | Zm00001eb019640 | 526 | 54 677.28 | 6.72 | 53 | 51 | 51.38 | 76.24 | -0.189 |
| ZmGRAS48 | Zm00001eb405080 | 235 | 25 649.61 | 6.55 | 27 | 25 | 48.23 | 99.66 | 0.063 |
| ZmGRAS49 | Zm00001eb002990 | 313 | 33 288.60 | 9.66 | 25 | 36 | 54.58 | 86.77 | 0.034 |
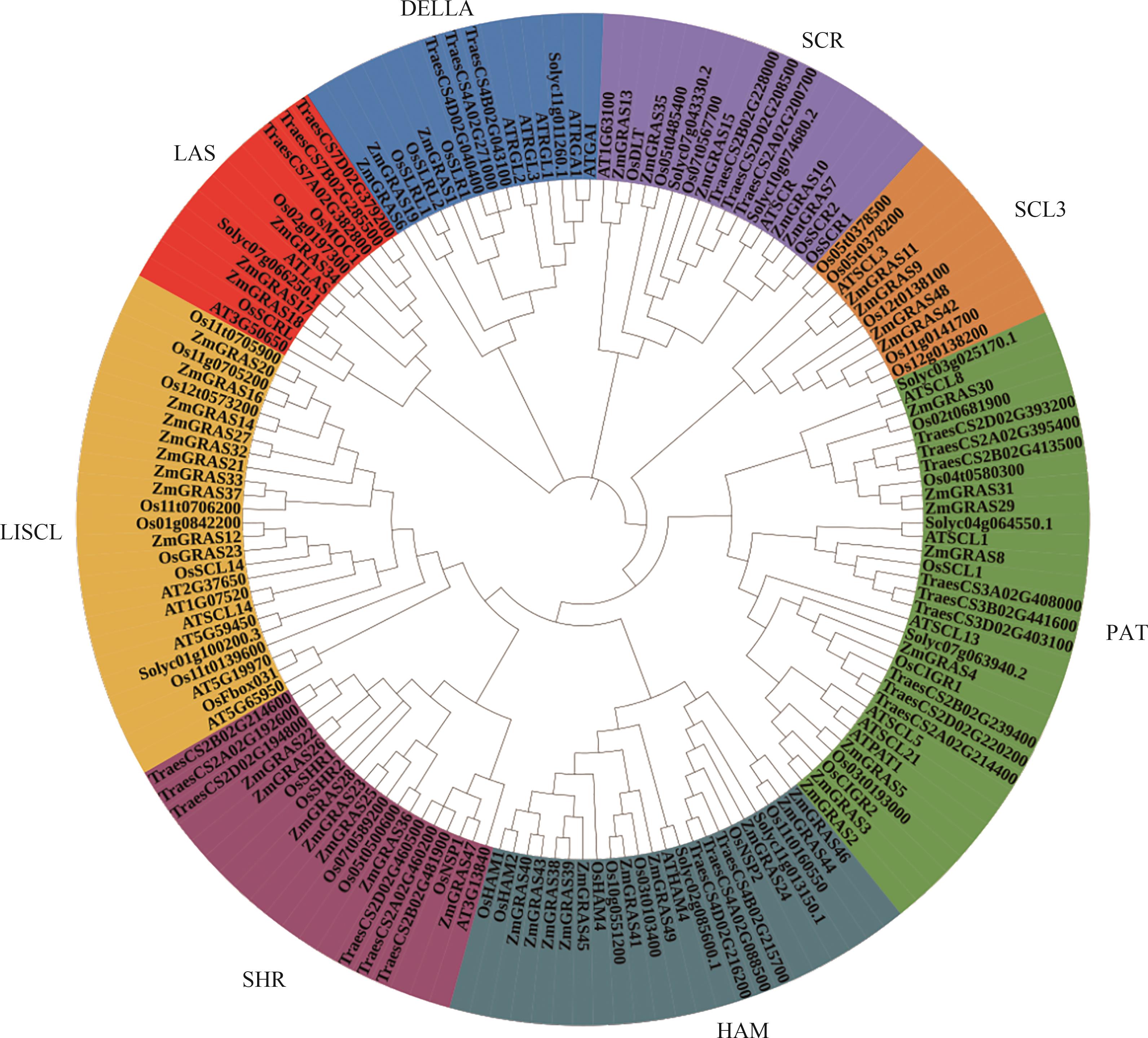
Fig. 2 Phylogenetic tree analysis of GRAS gene family in different speciesNote: Os—Oryza sativa; Zm—Zea mays; TresCS—Triticum aestivum Chinese Spring; At—Arabidopsis thaliana; Solyc—Solanum lycopersicum.

Fig. 5 Collinearity analysis of GRAS gene family members in maizeNote: The yellow-to-red scale bar on the right indicates the number of SNPs within 1 Mb domain.
| 1 | HOU S, ZHANG Q, CHEN J, et al.. Genome-wide identification and analysis of the GRAS transcription factor gene family in Theobroma cacao [J/OL]. Genes (Basel), 2022, 14(1):57 [2023-09-03]. . |
| 2 | LIU Y, HUANG W, XIAN Z, et al.. Overexpression of SIGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling [J/OL]. Front. Plant Sci., 2017, 8:1659 [2023-09-03]. . |
| 3 | WANG X, LI G, SUN Y, et al.. Genome-wide analysis and characterization of GRAS family in switchgrass [J]. Bioengineered, 2021, 12(1):6096-6114. |
| 4 | LI P, ZHANG B, SU T, et al.. BrLAS, a GRAS transcription factor from Brassica rapa, is involved in drought stress tolerance in transgenic arabidopsis [J/OL]. Front. Plant Sci., 2018, 9:1792 [2023-09-03]. . |
| 5 | LU X H, LIU W Q, XIANG C G, et al.. Genome-wide characterization of GRAS family and their potential roles in cold tolerance of cucumber (Cucumis sativus L.) [J/OL]. Int. J. Mol. Sci., 2020, 21(11):3857 [2023-09-03].. |
| 6 | BOLLE C. The role of GRAS proteins in plant signal transduction and development [J]. Planta, 2004, 218(5):683-692. |
| 7 | SUN X, JONES W T, RIKKERINK E H. GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling [J]. Biochem. J. 2012, 442(1):1-12. |
| 8 | 孔豆豆,毛成志,王蕾,等.大麦基因组GRAS基因家族的全基因组鉴定与表达分析 [J]. 分子植物育种, 2021, 19(1):22-33. |
| KONG D D, MAO C Z, WANG L, et al.. Genome-wide identification and phylogenetic analysis of the GRAS gene family in barley (Hordeum vulgare L.) [J]. Mol. Plant Breed., 2021, 19(1):22-33. | |
| 9 | 郑玲,闫晓曼.甜瓜GRAS家族全基因组的鉴定与表达分析[J].江苏农业科学, 2023, 51(11):53-59. |
| 10 | 张文慧,何光鑫,王子鑫,等.绿豆GRAS基因家族鉴定及其非生物胁迫下的表达模式分析[J].农业生物技术学报, 2022, 30(5):861-872. |
| ZHANG W H, HE G X, WANG Z X, et al.. Identification of GRAS gene family and its expression on pattern analysis under abiotic stress in Vigna radiata [J]. J. Agric. Biotechnol., 2022, 30(5):861-872. | |
| 11 | 王智兰,韩康妮,杜晓芬,等.谷子GRAS转录因子家族的全基因组鉴定、表达分析及标记开发 [J]. 核农学报, 2022, 36(9):1723-1737. |
| WANG Z L, HAN K N, DU X F, et al.. Identification, expression analysis and marker development of GRAS transcription factor in foxtail millet [J]. Acta Agric. Nucl. Sin., 2022, 36(9):1723-1737. | |
| 12 | TIAN C, WAN P, SUN S, et al.. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis [J]. Plant Mol. Biol., 2004, 54(4):519-532. |
| 13 | WANG T, YU T, FU J, et al.. Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance [J]. Front. Plant Sci., 2020, 11:604690 [2023-09-03]. . |
| 14 | 郭栋,宋雅菲,张佳阔,等.玉米CCCH基因家族鉴定及分析[J].中国农业科技导报, 2019, 21(8):19-27. |
| GUO D, SONG Y F, ZHANG J K, et al.. Identification and analysis of CCCH gene family in maize [J]. J. Agric. Sci. Technol., 2019, 21(8):19-27. | |
| 15 | HUSON D H, BRYANT D. Application of phylogenetic networks in evolutionary studies [J]. Mol. Biol. Evol., 2006, 23:254-267. |
| 16 | NAM J, MA H, NEI M. Antiquity and evolution of the MADSbox gene family controlling flower development in plants [J]. Mol. Biol. Evol., 2003, 20(9):1435-1447. |
| 17 | MOORE R C, PURUGGANAN M D. The evolutionary dynamics of plant duplicate genes [J]. Curr. Opin. Plant Biol., 2005, 8(2):122-128. |
| 18 | TANG H, BOWERS J E, WANG X, et al.. Synteny and collinearity in plant genomes [J]. Science, 2008, 320 (5875):486-488. |
| 19 | PENG J, CAROL P, RICHARDS D E, et al.. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses [J]. Genes Dev., 1997, 11:3194-3205. |
| 20 | SILVERSTONE A L, CIAMPAGLIO C N, SUN T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway [J]. Plant Cell, 1998, 10:155-169. |
| 21 | IKEDA A, UEGUCHI-TANAKA M, SONODA Y, et al.. Slender rice, a constitutive gibberellin response mutant.; is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8 [J]. Plant Cell, 2001, 13:999-1010. |
| 22 | STUURMAN J, JAGGI F, KUHLEMEIER C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells [J]. Genes Dev., 2002, 16:2213-2218. |
| 23 | MOROHASHI K, MINAMI M, TAKASE H, et al.. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression [J]. J. Biol. Chem., 2003, 278:20865-20873. |
| 24 | BOLLE C, KONCZ C, CHUA N H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction [J]. Genes Dev., 2000, 14:1269-1278. |
| 25 | YUAN Y, FANG L, KARUNGO S K, et al.. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis [J]. Plant Cell Rep., 2016, 35:655-666. |
| 26 | JI C, XU L N, LI Y J, et al.. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize [J]. Mol. Plant, 2022, 15(3):468-487. |
| 27 | LI Y, MA S, ZHAO Q Q, et al.. ZmGRAS11, transactivated by opaque2, positively regulates kernel size in maize [J]. J. Integr. Plant Biol., 2021, 63(12):2031-2037. |
| [1] | Wen DONG, Zehui JU, Zhenzhong WANG, Kangtai SUN. Project Establishment Analysis and Prospects of China’s “Forestry Germplasm Resources Cultivation and Quality Improvement” in the “14th Five-Year” National Key R&D Program [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 1-10. |
| [2] | Huaqiang TAN, Liping LI, Manman TIE, Jiaqin YANG, Xiaoyun ZHENG, Shaokun PAN, Youwan TANG. Mining and Analysis of Genes Related to Anthocyanin Degradation in Purple Pepper [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 79-91. |
| [3] | Zhien LIU, Yong HE, Zhicheng WANG, Xiaokang ZHAN, Tingbao WANG, Yaowei LIU, Zhihong TIAN. Identification and Bioinformatics Analysis of Growth Regulating Factor GRF Gene Family in Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 18-27. |
| [4] | Haitao XU, Hongzhen MA, Wenwen WANG, Wenxiang FAN, Bo XU, Jungang ZHANG, Haibin GUO, Youhua WANG. Research on Dynamic Development and Accumulated Temperature Model of Maize Plant Height and Stem Diameter Based on Effective Accumulated Temperature [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 187-201. |
| [5] | Zhining YANG, Xuke LU, Yapeng FAN, Yuping SUN, Xin YU, Liang WANG, Yongjian GAO, Gulijiayinashen Habuli, Gulishaxi Nasiyi, Wumuer Abudu, Hui HUANG, Menghao ZHANG, Lidong WANG, Xiao CHEN, Lei XIAO, Xinrui ZHANG, Shuai WANG, Xiugui CHEN, Junjuan WANG, Lixue GUO, Wenwei GAO, Wuwei YE. Response Mechanism of Cotton GhDMT7 Gene to 5-azacytidine [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 28-35. |
| [6] | Yongxu GUAN, Zhicheng SUN, Yan WANG, Yuan LI, Xiaoli SUN, Bowei JIA, Mingzhe SUN. Functional Analysis of Arabidopsis thalianaAtCHX19 Gene in Response to Salt-alkali Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 60-72. |
| [7] | Yiyang ZHANG, Tuoyu LIU, Yuan WANG, Jian TIAN, Feifei GUAN. Expression and Thermostability Modification of Alkaline Xylanase [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 73-79. |
| [8] | Xiang SUN, Liyang ZHANG, Jiying YIN, Kaiyi WANG, Meirong GUO, Heju HUAI. Problems and Countermeasures of Digital Transformation of Maize Seed Production Base [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 8-17. |
| [9] | Yi HU, Jie GONG, Wei ZHAO, Rong CHENG, Zhongyu LIU, Shiqing GAO, Yazhen YANG. Identification of PHY Gene Family in Wheat (Triticum aestivum L.)and Their Expression Analysis Under Heat Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 30-43. |
| [10] | Shichao CHEN, Ju WANG, Fuqiang GUO, Rui HAO, Jianping SHI. Effects of Different Water and Nitrogen Coupling on Physiological Indexes and Yield of Protein Mulberry [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 240-249. |
| [11] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [12] | Taotao MAO, Xiaoqiang ZHAO, Xiaodong BAI, Bin YU. Effect of Low Temperature Stress on Photosynthetic Performance, Antioxidant Enzyme System and Related Gene Expression in Maize Seedlings [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 49-60. |
| [13] | Lei LING, Huixin JIANG, Mingjing LI, Yajie YIN, Naiyu CHEN, Xiaoju ZHAO. Proteinome and Metabolome Combined to Analyze the Response Mechanism of Oat to Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 61-71. |
| [14] | Saisai HOU, Shanshan TONG, Pengqi WANG, Bingxue XIE, Ruifang ZHANG, Xinxin WANG. Effects of Biochar and Straw on Growth Characteristics and Nutrient Uptake of Different Crops [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 179-191. |
| [15] | Zihao WANG, Xue ZHOU, Donghan ZHANG, Hongyi LIANG, Yan WANG, Ziang ZHAO, Qing CHEN. Effect of Water-soluble Fertilizers Containing Humic-acids on Maize Seedlings Growth and Soil Properties [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 209-220. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号