




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (11): 76-86.DOI: 10.13304/j.nykjdb.2022.0341
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Huifeng KE1( ), Zhengwen SUN1, Guoning WANG1, Chengsheng MENG1, Liqiang WU2(
), Zhengwen SUN1, Guoning WANG1, Chengsheng MENG1, Liqiang WU2( )
)
Received:2022-04-23
Accepted:2022-05-28
Online:2022-11-15
Published:2022-11-29
Contact:
Liqiang WU
柯会锋1( ), 孙正文1, 王国宁1, 孟成生1, 吴立强2(
), 孙正文1, 王国宁1, 孟成生1, 吴立强2( )
)
通讯作者:
吴立强
作者简介:柯会锋E-mail: kehuifeng123@126.com;
基金资助:CLC Number:
Huifeng KE, Zhengwen SUN, Guoning WANG, Chengsheng MENG, Liqiang WU. Mining and Screening of Associated Markers and Candidate Genes Related to Seed Size and Shape in Cotton[J]. Journal of Agricultural Science and Technology, 2022, 24(11): 76-86.
柯会锋, 孙正文, 王国宁, 孟成生, 吴立强. 棉籽大小与形状关联标记发掘及候选基因筛选[J]. 中国农业科技导报, 2022, 24(11): 76-86.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2022.0341
| 性状 Trait | 年度 Year | 均值 Mean | 标准差 SD | 变异系数 CV/% | 最大值 Max. | 最小值 Min. | 偏度系数 Skew | 峰度系数 Kurt |
|---|---|---|---|---|---|---|---|---|
| 粒长 Seed length/mm | 2015 | 8.71 | 0.49 | 5.63 | 10.34 | 7.44 | 0.48 | 0.49 |
| 2017 | 8.20 | 0.45 | 5.49 | 10.54 | 7.09 | 0.59 | 1.52 | |
| 粒宽 Seed width/mm | 2015 | 4.74 | 0.23 | 4.85 | 5.46 | 4.06 | 0.16 | 0.30 |
| 2017 | 4.59 | 0.22 | 4.79 | 5.67 | 3.92 | 0.55 | 1.64 | |
| 籽粒长宽比 Ratio of seed length to width | 2015 | 1.85 | 0.08 | 4.32 | 2.11 | 1.65 | 0.41 | 0.50 |
| 2017 | 1.79 | 0.07 | 3.91 | 2.04 | 1.57 | 0.26 | 1.04 | |
| 籽粒面积 Seed area/mm2 | 2015 | 30.89 | 2.92 | 9.45 | 42.11 | 23.76 | 0.48 | 0.57 |
| 2017 | 28.69 | 2.74 | 9.55 | 45.16 | 22.38 | 0.89 | 3.07 | |
| 籽粒周长 Seed perimeter/mm | 2015 | 22.81 | 1.21 | 5.30 | 28.27 | 20.11 | 0.59 | 1.09 |
| 2017 | 21.43 | 1.10 | 5.13 | 27.32 | 18.96 | 0.65 | 1.71 |
Table 1 Genetic variation of seed related traits in cotton natural population
| 性状 Trait | 年度 Year | 均值 Mean | 标准差 SD | 变异系数 CV/% | 最大值 Max. | 最小值 Min. | 偏度系数 Skew | 峰度系数 Kurt |
|---|---|---|---|---|---|---|---|---|
| 粒长 Seed length/mm | 2015 | 8.71 | 0.49 | 5.63 | 10.34 | 7.44 | 0.48 | 0.49 |
| 2017 | 8.20 | 0.45 | 5.49 | 10.54 | 7.09 | 0.59 | 1.52 | |
| 粒宽 Seed width/mm | 2015 | 4.74 | 0.23 | 4.85 | 5.46 | 4.06 | 0.16 | 0.30 |
| 2017 | 4.59 | 0.22 | 4.79 | 5.67 | 3.92 | 0.55 | 1.64 | |
| 籽粒长宽比 Ratio of seed length to width | 2015 | 1.85 | 0.08 | 4.32 | 2.11 | 1.65 | 0.41 | 0.50 |
| 2017 | 1.79 | 0.07 | 3.91 | 2.04 | 1.57 | 0.26 | 1.04 | |
| 籽粒面积 Seed area/mm2 | 2015 | 30.89 | 2.92 | 9.45 | 42.11 | 23.76 | 0.48 | 0.57 |
| 2017 | 28.69 | 2.74 | 9.55 | 45.16 | 22.38 | 0.89 | 3.07 | |
| 籽粒周长 Seed perimeter/mm | 2015 | 22.81 | 1.21 | 5.30 | 28.27 | 20.11 | 0.59 | 1.09 |
| 2017 | 21.43 | 1.10 | 5.13 | 27.32 | 18.96 | 0.65 | 1.71 |
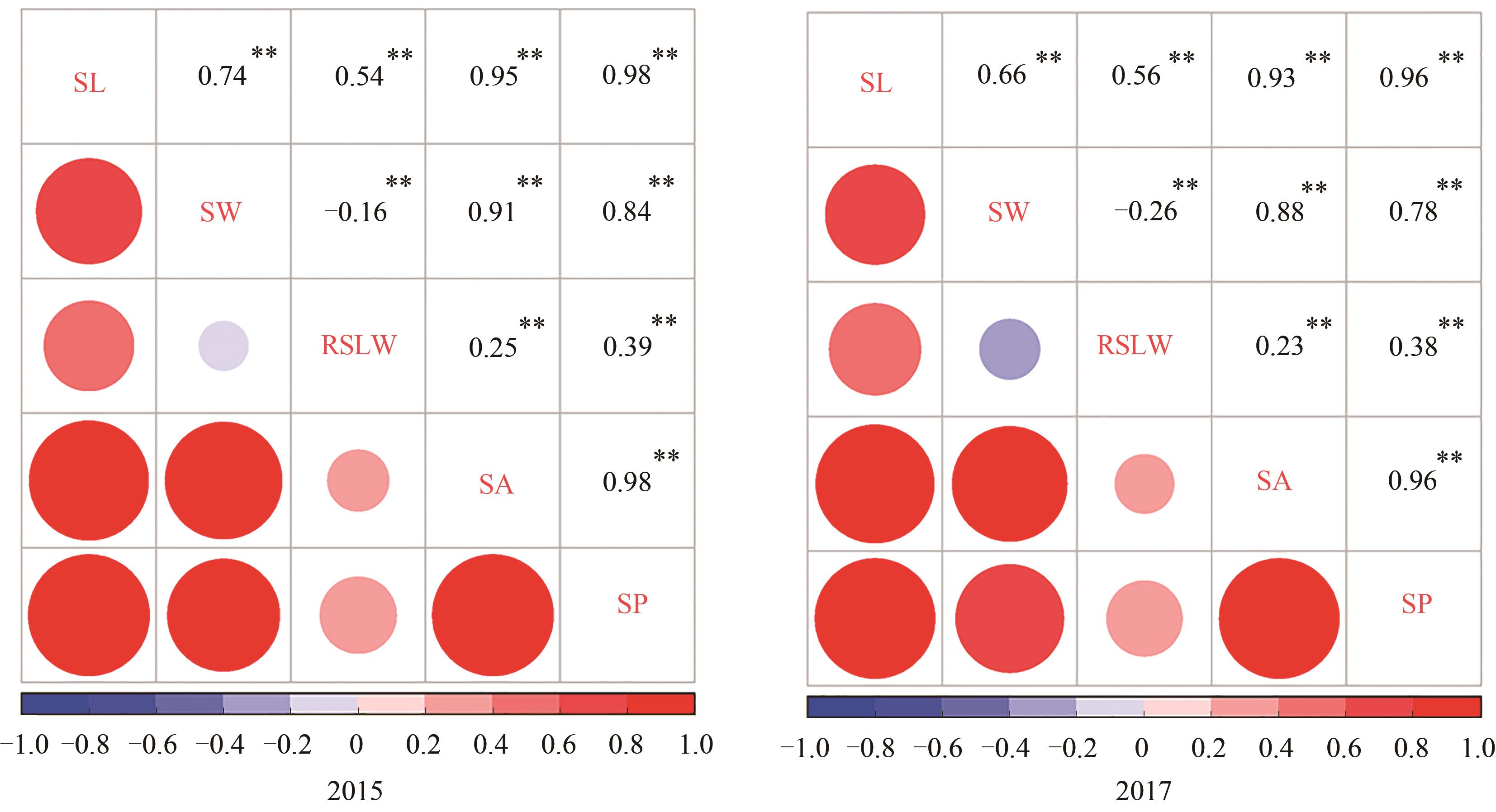
Fig. 2 Correlation of seed related traits in cotton natural populationNote: SL—Seed length; SW—Seed width; RSLW—Ratio of seed length to width; SA—Seed area; SP—Seed perimeter; ** indicates significant correlation at P<0.01 level.
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 5 | D05 | D05∶1496927 | 1 496 927 | T/C | RSLW | 2015, 2017 | 5.21, 5.34 |
| 6 | D05 | D05∶1512276 | 1 512 276 | T/A | RSLW | 2015, 2017 | 5.92, 5.25 |
| 7 | A04 | A04∶54398482 | 54 398 482 | C/A | SP | 2015, 2017 | 5.09, 5.32 |
| SA | 2017 | 5.22 | |||||
| 8# | A07 | A07∶70678488 | 70 678 488 | G/C | SL, SA,SP | 2015 | 6.89, 6.69, 6.72 |
| 9# | A07 | A07∶70678557 | 70 678 557 | A/G | SL,SA,SP | 2015 | 5.72, 5.35, 5.40 |
| 10# | A07 | A07∶70679600 | 70 679 600 | T/C | SL,SA,SP | 2015 | 5.18, 5.49, 5.27 |
| 11# | A07 | A07∶70679917 | 70 679 917 | C/T | SL,SA,SP | 2015 | 6.27, 5.87, 5.96 |
| 12# | A07 | A07∶70680268 | 70 680 268 | A/C | SL,SA,SP | 2015 | 6.14, 5.83, 5.84 |
| 13# | A07 | A07∶70680314 | 70 680 314 | A/G | SL,SA,SP | 2015 | 6.45, 6.38, 6.39 |
| 14# | A07 | A07∶70682150 | 70 682 150 | C/A | SL,SA,SP | 2015 | 5.87, 5.22, 5.51 |
| 15# | A07 | A07∶70682969 | 70 682 969 | T/C | SL,SA,SP | 2015 | 5.25, 5.19, 5.29 |
| 16# | A07 | A07∶70700642 | 70 700 642 | C/T | SL,SA,SP | 2015 | 5.56, 5.02, 5.16 |
| 17 | A07 | A07∶70706541 | 70 706 541 | C/G | SL,SA,SP | 2015 | 6.03, 5.21, 5.66 |
| 18 | A07 | A07∶72096450 | 72 096 450 | A/G | RSLW,SL,SP | 2015 | 5.10, 6.10, 5.12 |
| 19 | A07 | A07∶44855893 | 44 855 893 | G/A | SL,SA,SP | 2015 | 5.98, 6.06, 6.22 |
| 20 | A08 | A08∶28488249 | 28 488 249 | T/C | SL,SA,SP | 2015 | 5.17, 5.68, 5.64 |
| 21 | A08 | A08∶32230490 | 32 230 490 | T/C | SL,SA,SP | 2015 | 5.41, 5.48, 5.28 |
| 22 | A08 | A08∶97095613 | 97 095 613 | G/C | SL,SA,SP | 2015 | 5.23, 5.45, 5.76 |
| 23 | A01 | A01∶1540366 | 1 540 366 | C/T | SL,SA,SP | 2017 | 5.80, 5.25, 5.09 |
| 24 | A01 | A01∶12448791 | 12 448 791 | A/G | SL,SW,SA,SP | 2015 | 5.00, 5.89, 6.43, 5.85 |
| 25 | A02 | A02∶6005362 | 6 005 362 | C/T | SL,SA,SP | 2015 | 6.51, 5.03, 6.07 |
| 26 | A02 | A02∶10265596 | 10 265 596 | G/A | SL,SA,SP | 2017 | 6.20, 5.36, 5.90 |
| 27 | A05 | A05∶20705321 | 20 705 321 | C/G | SW,SA,SP | 2017 | 5.06, 5.04, 5.39 |
| 28 | A05 | A05∶37081549 | 37 081 549 | A/G | SL,SA,SP | 2017 | 6.63, 5.06, 6.17 |
| 29 | A06 | A06∶70904173 | 70 904 173 | A/G | SL,SA,SP | 2015 | 5.07, 5.05, 5.11 |
| 30 | D08 | D08∶34053083 | 34 053 083 | T/C | SL,SA,SP | 2017 | 5.90, 5.73, 5.74 |
Table 2 Associated SNP markers of seed size and shape in cotton natural population
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 5 | D05 | D05∶1496927 | 1 496 927 | T/C | RSLW | 2015, 2017 | 5.21, 5.34 |
| 6 | D05 | D05∶1512276 | 1 512 276 | T/A | RSLW | 2015, 2017 | 5.92, 5.25 |
| 7 | A04 | A04∶54398482 | 54 398 482 | C/A | SP | 2015, 2017 | 5.09, 5.32 |
| SA | 2017 | 5.22 | |||||
| 8# | A07 | A07∶70678488 | 70 678 488 | G/C | SL, SA,SP | 2015 | 6.89, 6.69, 6.72 |
| 9# | A07 | A07∶70678557 | 70 678 557 | A/G | SL,SA,SP | 2015 | 5.72, 5.35, 5.40 |
| 10# | A07 | A07∶70679600 | 70 679 600 | T/C | SL,SA,SP | 2015 | 5.18, 5.49, 5.27 |
| 11# | A07 | A07∶70679917 | 70 679 917 | C/T | SL,SA,SP | 2015 | 6.27, 5.87, 5.96 |
| 12# | A07 | A07∶70680268 | 70 680 268 | A/C | SL,SA,SP | 2015 | 6.14, 5.83, 5.84 |
| 13# | A07 | A07∶70680314 | 70 680 314 | A/G | SL,SA,SP | 2015 | 6.45, 6.38, 6.39 |
| 14# | A07 | A07∶70682150 | 70 682 150 | C/A | SL,SA,SP | 2015 | 5.87, 5.22, 5.51 |
| 15# | A07 | A07∶70682969 | 70 682 969 | T/C | SL,SA,SP | 2015 | 5.25, 5.19, 5.29 |
| 16# | A07 | A07∶70700642 | 70 700 642 | C/T | SL,SA,SP | 2015 | 5.56, 5.02, 5.16 |
| 17 | A07 | A07∶70706541 | 70 706 541 | C/G | SL,SA,SP | 2015 | 6.03, 5.21, 5.66 |
| 18 | A07 | A07∶72096450 | 72 096 450 | A/G | RSLW,SL,SP | 2015 | 5.10, 6.10, 5.12 |
| 19 | A07 | A07∶44855893 | 44 855 893 | G/A | SL,SA,SP | 2015 | 5.98, 6.06, 6.22 |
| 20 | A08 | A08∶28488249 | 28 488 249 | T/C | SL,SA,SP | 2015 | 5.17, 5.68, 5.64 |
| 21 | A08 | A08∶32230490 | 32 230 490 | T/C | SL,SA,SP | 2015 | 5.41, 5.48, 5.28 |
| 22 | A08 | A08∶97095613 | 97 095 613 | G/C | SL,SA,SP | 2015 | 5.23, 5.45, 5.76 |
| 23 | A01 | A01∶1540366 | 1 540 366 | C/T | SL,SA,SP | 2017 | 5.80, 5.25, 5.09 |
| 24 | A01 | A01∶12448791 | 12 448 791 | A/G | SL,SW,SA,SP | 2015 | 5.00, 5.89, 6.43, 5.85 |
| 25 | A02 | A02∶6005362 | 6 005 362 | C/T | SL,SA,SP | 2015 | 6.51, 5.03, 6.07 |
| 26 | A02 | A02∶10265596 | 10 265 596 | G/A | SL,SA,SP | 2017 | 6.20, 5.36, 5.90 |
| 27 | A05 | A05∶20705321 | 20 705 321 | C/G | SW,SA,SP | 2017 | 5.06, 5.04, 5.39 |
| 28 | A05 | A05∶37081549 | 37 081 549 | A/G | SL,SA,SP | 2017 | 6.63, 5.06, 6.17 |
| 29 | A06 | A06∶70904173 | 70 904 173 | A/G | SL,SA,SP | 2015 | 5.07, 5.05, 5.11 |
| 30 | D08 | D08∶34053083 | 34 053 083 | T/C | SL,SA,SP | 2017 | 5.90, 5.73, 5.74 |
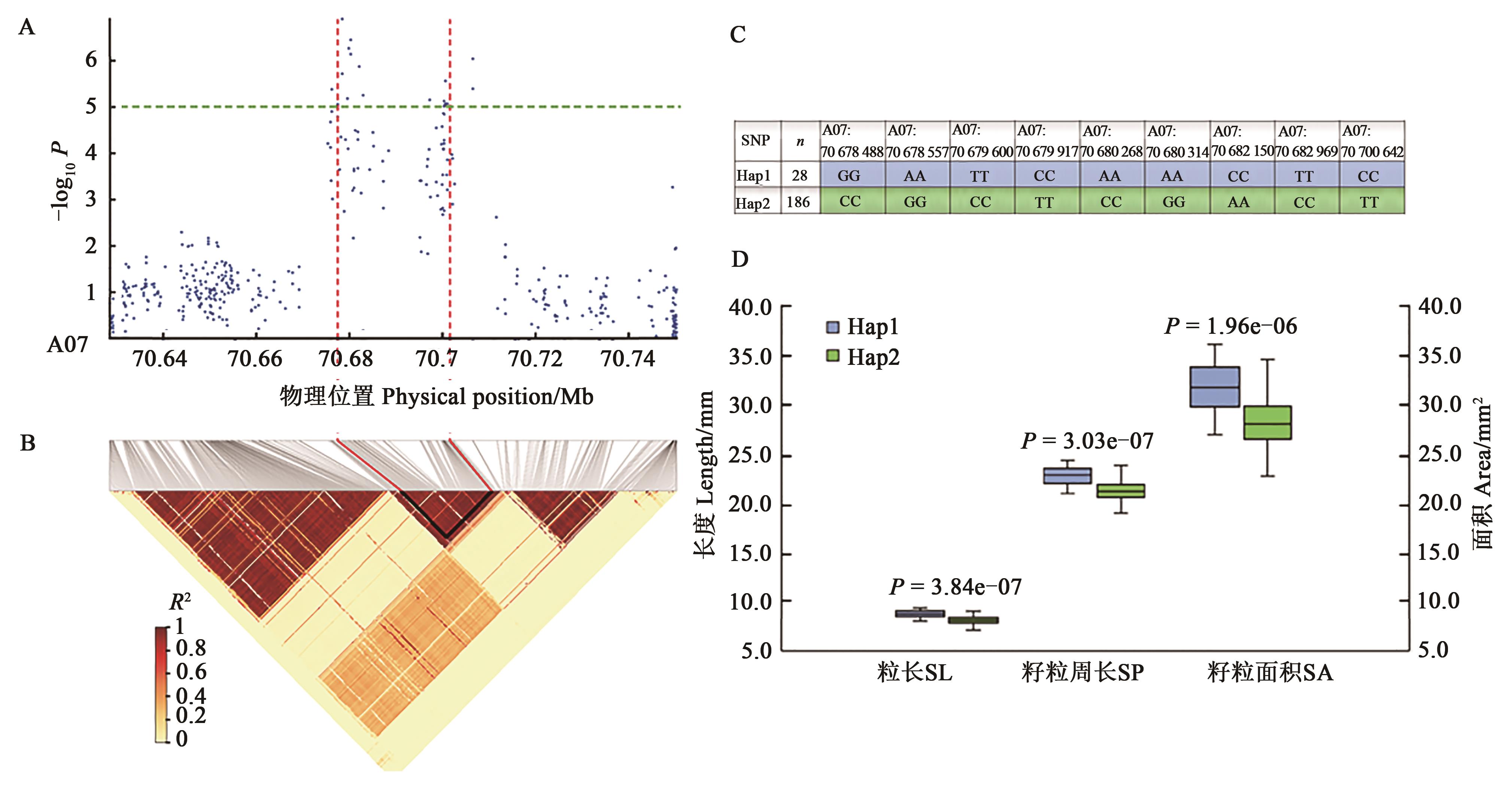
Fig. 3 Associated SNPs and haplotypes on chromosome A07 for cotton seed related traitsA: Associated SNPs on chromosome A07; B: LD analysis of chromosome A07; C: Haplotypes of associated SNPs; D: Different related traits between haplotypes
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 1 | D11 | D11∶50561153 | 50 561 153 | C/T | SW | 2015, 2017 | 5.32, 6.53 |
| SA | 2015, 2017 | 6.06, 6.45 | |||||
| SP | 2015, 2017 | 5.81, 5.76 | |||||
| 2 | D12 | D12∶56031535 | 56 031 535 | A/C | SL | 2015, 2017 | 5.85, 5.35 |
| SP | 2015, 2017 | 5.33, 5.46 | |||||
| SA | 2015 | 5.16 | |||||
| 3 | D05 | D05∶1490099 | 1 490 099 | G/A | RSLW | 2015, 2017 | 8.00, 6.83 |
| 4 | D05 | D05∶1490516 | 1 490 516 | G/T | RSLW | 2015, 2017 | 5.63, 5.36 |
Table 2 Associated SNP markers of seed size and shape in cotton natural population
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 1 | D11 | D11∶50561153 | 50 561 153 | C/T | SW | 2015, 2017 | 5.32, 6.53 |
| SA | 2015, 2017 | 6.06, 6.45 | |||||
| SP | 2015, 2017 | 5.81, 5.76 | |||||
| 2 | D12 | D12∶56031535 | 56 031 535 | A/C | SL | 2015, 2017 | 5.85, 5.35 |
| SP | 2015, 2017 | 5.33, 5.46 | |||||
| SA | 2015 | 5.16 | |||||
| 3 | D05 | D05∶1490099 | 1 490 099 | G/A | RSLW | 2015, 2017 | 8.00, 6.83 |
| 4 | D05 | D05∶1490516 | 1 490 516 | G/T | RSLW | 2015, 2017 | 5.63, 5.36 |
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation | 表达量增加的时期 Stage with increasing expression |
|---|---|---|---|
| D05 | Gh_D05G0142 | 铜离子结合,电子载体 Copper ion binding, electron carrier | 后期 Later stage |
| Gh_D05G0144 | 植物特异转录因子,YABBY家族蛋白 Plant-specific transcription factor, YABBY family protein | 后期Later stage | |
| Gh_D05G0145 | 核孔蛋白相关 Nucleoporin-related | 后期Later stage | |
| Gh_D05G0146 | TPR超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | 全期 Whole stage | |
| Gh_D05G0147 | AT1G80133 | 全期Whole stage | |
| Gh_D05G0148 | EBF蛋白 EIN3-binding F box protein | 全期Whole stage | |
| Gh_D05G0149 | AT5G25360 | 前期Earlier stage | |
| Gh_D05G0150 | AT3G15760 | 前期Earlier stage | |
| Gh_D05G0153 | 含SPX结构域基因 SPX domain gene | 全期Whole stage | |
| Gh_D05G0154 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 前期Earlier stage | |
| Gh_D05G0155 | 核糖核酸酶H超家族蛋白 Ribonuclease H-like superfamily protein | 后期Later stage | |
| A07 | Gh_A07G1731 | AT3G13130 | 后期Later stage |
| Gh_A07G1732 | AT3G13130 | 全期Whole stage | |
| A08 | Gh_A08G0768 | 蛋白激酶超家族蛋白 Protein kinase superfamily protein | 全期Whole stage |
| Gh_A08G0782 | 海藻糖磷酸酶/合酶 Trehalose phosphatase/synthase | 前期Earlier stage | |
| A01 | Gh_A01G0158 | 富亮氨酸跨膜蛋白激酶 Leucine-rich repeat transmembrane protein kinase | 全期Whole stage |
| Gh_A01G0160 | 衰老相关基因 Senescence-associated gene | 后期Later stage | |
| Gh_A01G0695 | 含SET结构域蛋白 SET domain group 40 | 全期Whole stage | |
| A02 | Gh_A02G0442 | RING/U-box 超家族蛋白 RING/U-box superfamily protein | 全期Whole stage |
| Gh_A02G0645 | Gigantea蛋白 Gigantea protein | 后期Later stage | |
| A05 | Gh_A05G2586 | 类PsbP蛋白 PsbP-like protein | 后期Later stage |
Table 3 Candidate genes related to the seed size and shape in cotton
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation | 表达量增加的时期 Stage with increasing expression |
|---|---|---|---|
| D05 | Gh_D05G0142 | 铜离子结合,电子载体 Copper ion binding, electron carrier | 后期 Later stage |
| Gh_D05G0144 | 植物特异转录因子,YABBY家族蛋白 Plant-specific transcription factor, YABBY family protein | 后期Later stage | |
| Gh_D05G0145 | 核孔蛋白相关 Nucleoporin-related | 后期Later stage | |
| Gh_D05G0146 | TPR超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | 全期 Whole stage | |
| Gh_D05G0147 | AT1G80133 | 全期Whole stage | |
| Gh_D05G0148 | EBF蛋白 EIN3-binding F box protein | 全期Whole stage | |
| Gh_D05G0149 | AT5G25360 | 前期Earlier stage | |
| Gh_D05G0150 | AT3G15760 | 前期Earlier stage | |
| Gh_D05G0153 | 含SPX结构域基因 SPX domain gene | 全期Whole stage | |
| Gh_D05G0154 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 前期Earlier stage | |
| Gh_D05G0155 | 核糖核酸酶H超家族蛋白 Ribonuclease H-like superfamily protein | 后期Later stage | |
| A07 | Gh_A07G1731 | AT3G13130 | 后期Later stage |
| Gh_A07G1732 | AT3G13130 | 全期Whole stage | |
| A08 | Gh_A08G0768 | 蛋白激酶超家族蛋白 Protein kinase superfamily protein | 全期Whole stage |
| Gh_A08G0782 | 海藻糖磷酸酶/合酶 Trehalose phosphatase/synthase | 前期Earlier stage | |
| A01 | Gh_A01G0158 | 富亮氨酸跨膜蛋白激酶 Leucine-rich repeat transmembrane protein kinase | 全期Whole stage |
| Gh_A01G0160 | 衰老相关基因 Senescence-associated gene | 后期Later stage | |
| Gh_A01G0695 | 含SET结构域蛋白 SET domain group 40 | 全期Whole stage | |
| A02 | Gh_A02G0442 | RING/U-box 超家族蛋白 RING/U-box superfamily protein | 全期Whole stage |
| Gh_A02G0645 | Gigantea蛋白 Gigantea protein | 后期Later stage | |
| A05 | Gh_A05G2586 | 类PsbP蛋白 PsbP-like protein | 后期Later stage |
| 1 | NIE X H, TU J L, WANG B, et al.. A BIL population derived from G . hirsutumG.barbadense provides a resource for cotton genetics and breeding [J/OL]. PLoS One, 2015, 10(10): e01410641 [2022-04-19]. . |
| 2 | LIU H Y, QUAMPAH A, CHEN J H, et al.. QTL mapping with different genetic systems for nine nonessential amino acids of cottonseeds [J]. Mol. Genet. Genomics, 2017, 292(10): 671-684. |
| 3 | SHANG L G, ABDUWELI A, WANG Y M, et al.. Genetic analysis and QTL mapping of oil content and seed index using two recombinant inbred lines and two backcross populations in upland cotton [J]. Plant Breeding, 2016, 135(2): 224-231. |
| 4 | 钱玉源,刘祎,张曦,等.棉花种子质量突变体ims-15及其近等基因系种子发育期转录组分析[J].华北农学报, 2021, 36 (3): 50-59. |
| QIAN Y Y, LIU Y, ZHANG X, et al.. Transcriptome analysis of cotton seed weight mutant ims-15 and its near isogenic line during seed development [J]. Acta Agric. Boreali-Sin., 2021, 36 (3): 50-59. | |
| 5 | 赵娟.调控内源生长素和细胞分裂素水平实现棉花种子纤维产量和品质的同步改良[D].重庆:西南大学, 2014. |
| ZHAO J. Manipulation of auxin and cytokinin level in cotton concurrently increases fiber and seed yield [D]. Chongqing: Southwest University, 2014. | |
| 6 | 覃珊.胚中特异性下调细胞分裂素氧化酶基因(GhCKX)促进棉花种子发育[D].重庆:西南大学, 2009. |
| QIN S. Ovule-specific down-regulation of GhCKX gene promoted seed development of cotton (Gossypium hirsutum L.) [D]. Chongqing: Southwest University, 2009. | |
| 7 | 王俊娟,叶武威,樊伟莉,等.不同基因型棉花种子活力的研究[J].种子科技, 2009, 27(9): 19-20. |
| WANG J J, YE, W W, FAN W L, et al.. Study on vitality of different genotypes cotton seed [J]. Seed Sci. Technol., 2009, 27(9): 19-20. | |
| 8 | ZHANG D, LI J P, COMPTON R O, et al.. Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae) [J]. G3: Genes Genom. Genet., 2015, 5(6): 1117-1128. |
| 9 | LI L, LI X H, LI L L, et al.. QTL identification and epistatic effect analysis of seed size and weight-related traits in Zea mays L [J/OL]. Mol. Breeding, 2019, 39(5): 67 [2022-04-20]. . |
| 10 | WILLIAMS K, SORRELLS M E. Three-dimensional seed size and shape QTL in hexaploid wheat (Triticum aestivum L.) populations [J]. Crop Sci., 2014, 54(1): 98-110. |
| 11 | CUI B F, CHEN L, YANG Y Q, et al.. Genetic analysis and map-based delimitation of a major locus qSS3 for seed size in soybean [J]. Plant Breeding, 2020, 139(2): 1145-1157. |
| 12 | HINA A, CAO Y C, SONG S Y, et al.. High-resolution mapping in two RIL populations refines major "QTL hotspot" regions for seed size and shape in soybean (Glycine max L.) [J/OL]. Int. J. Mol. Sci., 2020, 21(3): 1040 [2022-04-20]. . |
| 13 | GERAVANDI M, CHEGHAMIRZA K, FARSHADFAR E, et al.. QTL analysis of seed size and yield-related traits in an inter-gene pool population of common bean (Phaseolus vulgaris L.) [J/OL]. Scientia Hortic., 2020, 274(15): 109678 [2022-05-20]. . |
| 14 | SONG X J, HUANG W, SHI M, et al.. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase [J]. Nat. Genet., 2007, 39(5): 623-630. |
| 15 | 程瑞如.小麦粒形的分子遗传分析及穗长基因HL1的精细定位[D].南京:南京农业大学, 2018. |
| CHENG R R. Molecular genetic analysis of wheat kernel dimensions and fine mapping of spike length QTL HLl [D]. Nanjing: Nanjing Agricultural University, 2018. | |
| 16 | 耿庆河,王兰芬,武晶,等.普通菜豆籽粒大小与形状的QTL定位[J].作物学报,2017, 43(8): 1149-1160. |
| GENG Q H, WANG L F, WU J, et al.. QTL mapping for seed size and shape in common bean [J]. Acta Agron. Sin., 2017, 43(8): 1149-1160. | |
| 17 | 秦伟伟,李永祥,李春辉,等.基于高密度遗传图谱的玉米籽粒性状QTL定位[J].作物学报, 2015, 41(10): 1510-1518. |
| QIN W W, LI Y X, LI C H, et al.. QTL mapping for kernel related traits based on a high-density genetic map [J]. Acta Agron. Sin., 2015, 41(10): 1510-1518. | |
| 18 | 耿青青.大豆籽粒大小与形状性状的全基因组关联分析和DistortedMap软件包的研制[D].南京:南京农业大学, 2014. |
| GENG Q Q. Genome-wide association studies of seed size and shape traits in soybean and development of distortedmap software [D]. Nanjing: Nanjing Agricultural University, 2014. | |
| 19 | 曾雅青.棉花种子活力的基因型差异及生态点影响[D].南京:南京农业大学, 2014. |
| ZENG Y Q. Genotype difference and ecological environmental impact in cotton seed vigor [D]. Nanjing: Nanjing Agricultural University, 2014. | |
| 20 | 陈建华,杨继良,杨伟华,等.棉花种子活力及其影响因素探讨[J].棉花学报, 1995, 7(3): 134-140. |
| CHEN J H, YANG J L, YANG W H, et al.. The study of cottonseed vigor and influence factors [J]. Acta Gossypii Sin., 1995, 7(3): 134-140. | |
| 21 | MA Z Y, HE S P, WANG X F, et al.. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield [J]. Nat. Genet., 2018, 50(6): 803-813. |
| 22 | GU Q S, KE H F, SUN Z W, et al.. Genetic variation associated with the shoot biomass of upland cotton seedlings under contrasting phosphate supplies [J/OL]. Mol. Breeding, 2020, 40(8): 80 [2022-04-20]. . |
| 23 | FANG Y X, ZHANG X Q, ZHANG X, et al.. A high-density genetic linkage map of SLAFs and QTL analysis of grain size and weight in barley (Hordeum vulgare L.) [J/OL]. Front. Plant Sci., 2020, 11: 620922 [2022-04-20]. . |
| 24 | LEI L, WANG L F, WANG S M, et al.. Marker-trait association analysis of seed traits in accessions of common bean (Phaseolus vulgaris L.) in China [J/OL]. Front. Genet., 2020, 11: 698 [2022-04-20]. . |
| 25 | ZHANG T Z, HU Y, JIANG W K, et al.. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement [J]. Nat. Biotechnol., 2015, 33(5): 531-537. |
| 26 | GUO X K, ZHANG Y, TU Y, et al.. Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato [J]. Plant Sci., 2018, 272: 131-141. |
| 27 | 陈秀玲,刘思源,孙婷,等.番茄YABBY基因家族的生物信息学分析[J].东北农业大学学报, 2017, 48(10): 11-19. |
| CHEN X L, LIU S Y, SUN T, et al.. Bioinformatics analysis on YABBY gene family in tomato [J]. J. Northeast Agric. Univ., 2017, 48(10): 11-19. | |
| 28 | ZHANG M, WANG C P, LIN Q F, et al.. A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis [J]. Plant J., 2015, 83(4): 582-599. |
| 29 | WEI K F, LI Y X. Functional genomics of the protein kinase superfamily from wheat [J/OL]. Mol. Breeding, 2019, 39: 141 [2022-04-20]. . |
| 30 | WANG W W, SUN Y, YANG P, et al.. A high density SLAF-seq SNP genetic map and QTL for seed size, oil and protein content in upland cotton [J/OL]. BMC Genomics, 2019, 20(1): 599 [2022-04-20]. . |
| 31 | FINET C, FLOYD S K, CONWAY S J, et al.. Evolution of the YABBY gene family in seed plants [J]. Evol. Dev., 2016, 18(2): 116-126. |
| 32 | SUN L, WEI Y Q, WU K H, et al.. Restriction of iron loading into developing seeds by a YABBY transcription factor safeguards successful reproduction in Arabidopsis [J]. Mol. Plant, 2021, 14(10): 1624-1639. |
| [1] | Hao JIA, Hongzhe WANG, Zhengwen SUN, Qishen GU, Dongmei ZHANG, Xingyi WANG, Yan ZHANG, Huaiyu LU, Zhiying MA, Xingfen WANG. Genome-wide Identification of VOZ Genes Family in Cotton and Study on Salt Tolerance Function of GhVOZ1 [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 58-68. |
| [2] | Guiyuan ZHAO, Yongqiang WANG, Jianguang LIU, Zhao GENG, Hanshuang ZHANG, Liqiang WU, Xingfen WANG, Guiyin ZHANG. Effect of Exogenous Gene Insertion Site on Bt Protein Content in Insect-resistant Cotton [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 44-53. |
| [3] | Yixin CHEN, Xiubo YANG, Shijun TIAN, Cong WANG, Zhiying BAI, Cundong LI, Ke ZHANG. Response of GhCOMT28 to Drought Stress in Gossypium hirsutum [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 45-56. |
| [4] | Zhiduo DONG, Qiuping FU, Jian HUANG, Tong QI, Yanbo FU, Kuerban Kaisaier. Analysis of Salt Tolerance Capacity of Xinjiang Cotton Guring Germination [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 57-67. |
| [5] | Zicheng PENG, Hongli DU, Ming WANG, Fenghua ZHANG, Haichang YANG. Research on AMF Regulation of Cotton Growth and Ion Balance Under Salt Alkali Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(2): 33-41. |
| [6] | Songjiang DUAN, Haoran HU, Chengjie ZHANG, Wei SUN, Yifan WU, Rensong GUO, Jusong ZHANG. Differences in Nitrogen Efficiency of Different Genotypes of Island Cotton and Their Effects on Photosynthetic Characteristics and Yield [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 61-71. |
| [7] | Huiting WENG, Haiyang LIU, Huiming GUO, Hongmei CHENG, Jun LI, Chao ZHANG, Xiaofeng SU. Preliminary Function Analysis of GhERF020 Gene in Response to Verticillium Wilt in Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 112-121. |
| [8] | Ronghua WEI, Ming YIN, Wensheng WANG, Yanru CUI. Discovering of QTLs and Candidate Genes Related to Rice Heading Period Traits Based on BSA-seq [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 12-24. |
| [9] | Ziqin LI, Jiaqiang WANG, Zhen LI, Deqiu ZOU, Xiaogong ZHANG, Xiaoyu LUO, Weiyang LIU. Estimation of Chlorophyll Density of Cotton Leaves Based on Spectral Index [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 103-111. |
| [10] | Yukun QIN, Junying CHEN, Lijuan ZHANG. Response of Dry Matter Accumulation Characteristics and Yield of Cotton in North Jiangxi Cotton Region to Nitrogen Reduction Measures [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 191-199. |
| [11] | Ling LIN, Yujie ZHU, Lei FENG, Guangmu TANG, Yunshu ZHANG, Wanli XU. Effects of Aged Cotton Straw Biochars on Soil Properties and Nitrogen Utilization of Wheat [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 184-191. |
| [12] | Jiangbo LI, Wenju GAO, Xiaodong YUN, Jieyin ZHAO, Shiwei GENG, Chunbin HAN, Quanjia CHEN, Qin CHEN. Effects of Different Water Stress Treatments on Core Germplasm Resources of Upland Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 26-39. |
| [13] | Lihua LI, Zhengwen SUN, Huifeng KE, Qishen GU, Liqiang WU, Yan ZHANG, Guiyin ZHANG, Xingfen WANG. Development and Effect Evaluation of KASP Markers for Fiber Strength in Gossypium hirsutum L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 46-55. |
| [14] | Menghua ZHAI, Minghui SUN, Xuerui LI, Xinlong XU, Haizhou GAO, Jusong ZHANG. Effects of DPC on Plant Type Shaping of Cotton Under Different Plant Spacing Configurations [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 145-156. |
| [15] | Zhen CHENG, Jianlong NIU, Yuting MA, weiyang LIU, Xuewei JIANG, Xueqi LIANG, Hongqiang DONG. Dynamic Changes of Cotton Phenological Stages in Alar Reclamation Area of Southern Xinjiang from 1990 to 2020 [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 206-214. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号