




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (6): 36-46.DOI: 10.13304/j.nykjdb.2021.0806
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Nan WU( ), Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG(
), Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG( )
)
Received:2021-09-13
Accepted:2021-11-22
Online:2022-06-15
Published:2022-06-21
Contact:
Xingfen WANG
吴楠( ), 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬(
), 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬( )
)
通讯作者:
王省芬
作者简介:吴楠 E-mail:wn2013@126.com;
基金资助:CLC Number:
Nan WU, Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG. Overexpression of a Cotton Glucuronokinase Gene GbGlcAK Promotes Cell Elongation in Arabidopsis thaliana[J]. Journal of Agricultural Science and Technology, 2022, 24(6): 36-46.
吴楠, 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬. 过表达棉花葡萄糖醛酸激酶基因GbGlcAK促进拟南芥细胞伸长[J]. 中国农业科技导报, 2022, 24(6): 36-46.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2021.0806
引物名称 Primer names | 登录号 Accession No. | 引物序列 Primer sequences (5’-3’) | 用途 Purpose |
|---|---|---|---|
| GlcAK-F | — | TGGTATTGGCTTGGTGCAGT | 基因克隆 Gene cloning |
| GlcAK-R | CAAAAACAATCCCTAAAATGCTCTT | ||
| sGbGlcAK-R | — | GGTGGTACCCTGCTTAGACAATGT | 亚细胞定位Subcellular location |
| GbGlcAK-F | — | GGCGTCTAGAATGGATCAAAATATG | 亚细胞定位和载体构建 Subcellular location and vector construction |
| GbGlcAK-R | GGTACCCGCCTACTTAGACAATG | 载体构建 Vector construction | |
| qGbGlcAK-F | — | CATCATCAACCCTCACCCCATTC | 拟南芥实时定量PCR Real-time PCR for A. thaliana |
| qGbGlcAK-R | GCCGCATACAATAGCACTGGACC | ||
| qAtGlcAK-F | AT3G01640 | GGATAGAGCATCGGTCCTTCGC | |
| qAtGlcAK-R | CGCCATTAGCAACCTTACCCCA | ||
| qAtMIPS-F | AT4G39800 | AAGGAGAAGAATAAGGTGGATAAGGTT | |
| qAtMIPS-R | GCAATCGCATAAAGTGTTGAAGG | ||
| qAtIMP-F | AT3G02870 | GACAGAGACTGATAAAGGATGTGAAGA | |
| qAtIMP-R | AGGGAACCCGTGAACGAAAT | ||
| qAtMIOX-F | AT4G26260 | CGAAGCCATCCGCAAAGATTACC | |
| qAtMIOX-R | CCAACAACAGCCCATTGAGGAAGTC | ||
| qAtPGM-F | AT1G23190 | GCTACCTACGGTCGTCACTATTACACTCG | |
| qAtPGM-R | GTAACGGATTCCCTGGTGCTTCG | ||
| qAtUGP-F | AT5G17310 | GCCCAGCAAGGGAAAGACCG | |
| qAtUGP-R | CGATGGCACCCAAGTTGTCTGAAT | ||
| qAtUGD-F | AT5G15490 | GATTGCGGTTCTCGGCTTCG | |
| qAtUGD-R | TGCTTCACAGTGGTGGGGCTC | ||
| qAtUXS-F | AT5G59290 | TGAGAAGAATGAGGTGGTTGTTGC | |
| qAtUXS-R | GGTTGTATTTGTAGAAGATAGGAGAGGC | ||
| qAtGAE-F | AT3G23820 | GCGACGGCGGATACAAGCA | |
| qAtGAE-R | GATGGCGAAGATGAGGACGAGG | ||
| qAtUB5-F | AT3G62250 | CCTCGCCGACTACAACATCCAG | |
| qAtUB5-R | CTTCTTCCTCTTCTTAGCACCACCA |
Table 1 PCR and qPCR primers used in this study
引物名称 Primer names | 登录号 Accession No. | 引物序列 Primer sequences (5’-3’) | 用途 Purpose |
|---|---|---|---|
| GlcAK-F | — | TGGTATTGGCTTGGTGCAGT | 基因克隆 Gene cloning |
| GlcAK-R | CAAAAACAATCCCTAAAATGCTCTT | ||
| sGbGlcAK-R | — | GGTGGTACCCTGCTTAGACAATGT | 亚细胞定位Subcellular location |
| GbGlcAK-F | — | GGCGTCTAGAATGGATCAAAATATG | 亚细胞定位和载体构建 Subcellular location and vector construction |
| GbGlcAK-R | GGTACCCGCCTACTTAGACAATG | 载体构建 Vector construction | |
| qGbGlcAK-F | — | CATCATCAACCCTCACCCCATTC | 拟南芥实时定量PCR Real-time PCR for A. thaliana |
| qGbGlcAK-R | GCCGCATACAATAGCACTGGACC | ||
| qAtGlcAK-F | AT3G01640 | GGATAGAGCATCGGTCCTTCGC | |
| qAtGlcAK-R | CGCCATTAGCAACCTTACCCCA | ||
| qAtMIPS-F | AT4G39800 | AAGGAGAAGAATAAGGTGGATAAGGTT | |
| qAtMIPS-R | GCAATCGCATAAAGTGTTGAAGG | ||
| qAtIMP-F | AT3G02870 | GACAGAGACTGATAAAGGATGTGAAGA | |
| qAtIMP-R | AGGGAACCCGTGAACGAAAT | ||
| qAtMIOX-F | AT4G26260 | CGAAGCCATCCGCAAAGATTACC | |
| qAtMIOX-R | CCAACAACAGCCCATTGAGGAAGTC | ||
| qAtPGM-F | AT1G23190 | GCTACCTACGGTCGTCACTATTACACTCG | |
| qAtPGM-R | GTAACGGATTCCCTGGTGCTTCG | ||
| qAtUGP-F | AT5G17310 | GCCCAGCAAGGGAAAGACCG | |
| qAtUGP-R | CGATGGCACCCAAGTTGTCTGAAT | ||
| qAtUGD-F | AT5G15490 | GATTGCGGTTCTCGGCTTCG | |
| qAtUGD-R | TGCTTCACAGTGGTGGGGCTC | ||
| qAtUXS-F | AT5G59290 | TGAGAAGAATGAGGTGGTTGTTGC | |
| qAtUXS-R | GGTTGTATTTGTAGAAGATAGGAGAGGC | ||
| qAtGAE-F | AT3G23820 | GCGACGGCGGATACAAGCA | |
| qAtGAE-R | GATGGCGAAGATGAGGACGAGG | ||
| qAtUB5-F | AT3G62250 | CCTCGCCGACTACAACATCCAG | |
| qAtUB5-R | CTTCTTCCTCTTCTTAGCACCACCA |
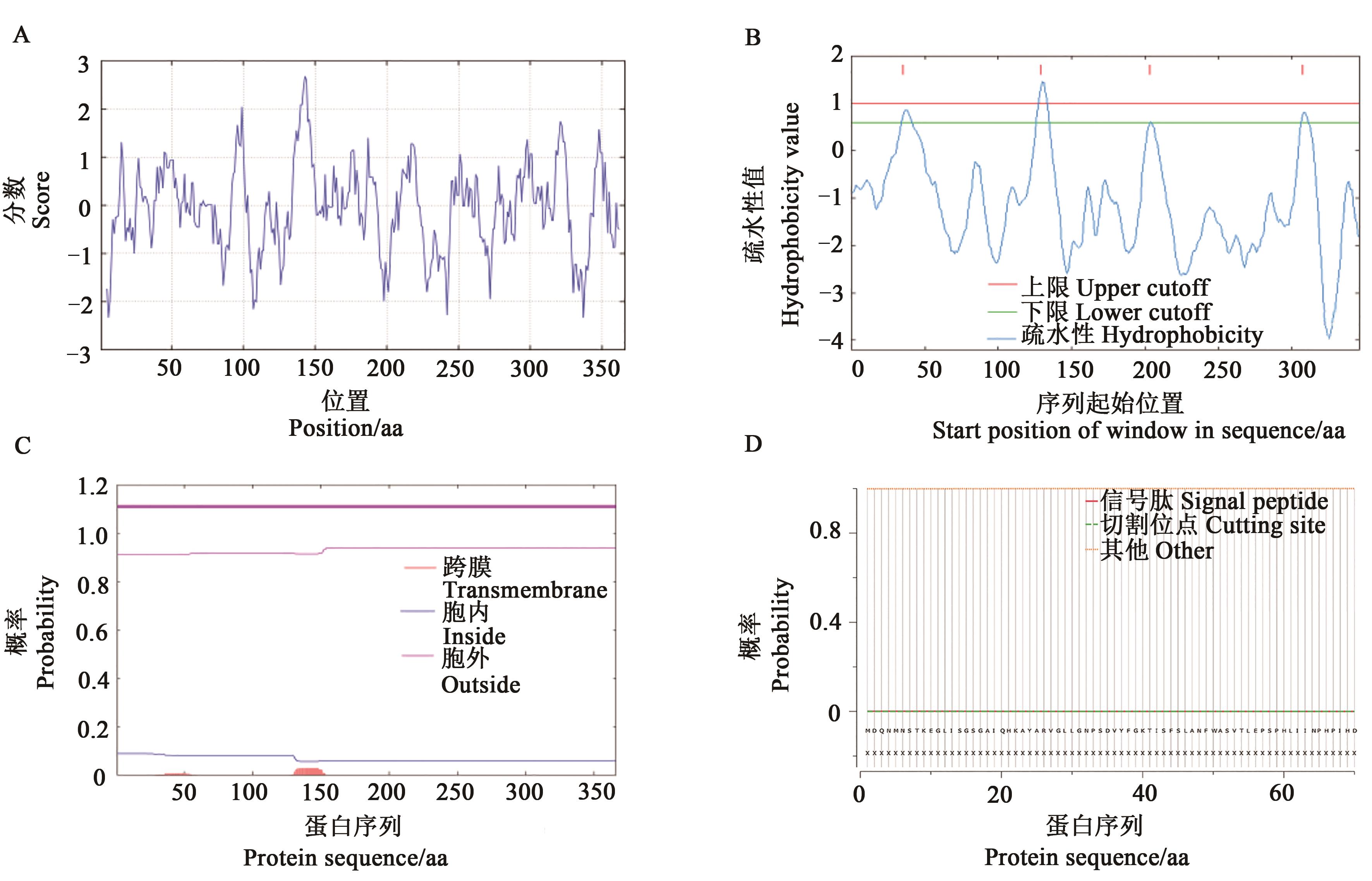
Fig. 1 Characterization of GbGlcAK proteinA: Hydrophilic/hydrophobic distribution curve of the GbGlcAK peptide analyzed by ProtScale; B, C: Transmembrane region prediction of the GbGlcAK amino acid sequence by TOPpred2 and TMHMM 2.0; D: Signal peptide analysis of the GbGlcAK protein

Fig.2 Evolutionary analysis of GbGlcAKNote: Rc—Ricinus communis; Pt—Populus trichocarpa; Vv—Vitis vinifera; At—Arabidopsis thaliana; Gm—Glycine max; Os—Oryza sativa; Zm—Zea mays; Sb—Sorghum bicolor.
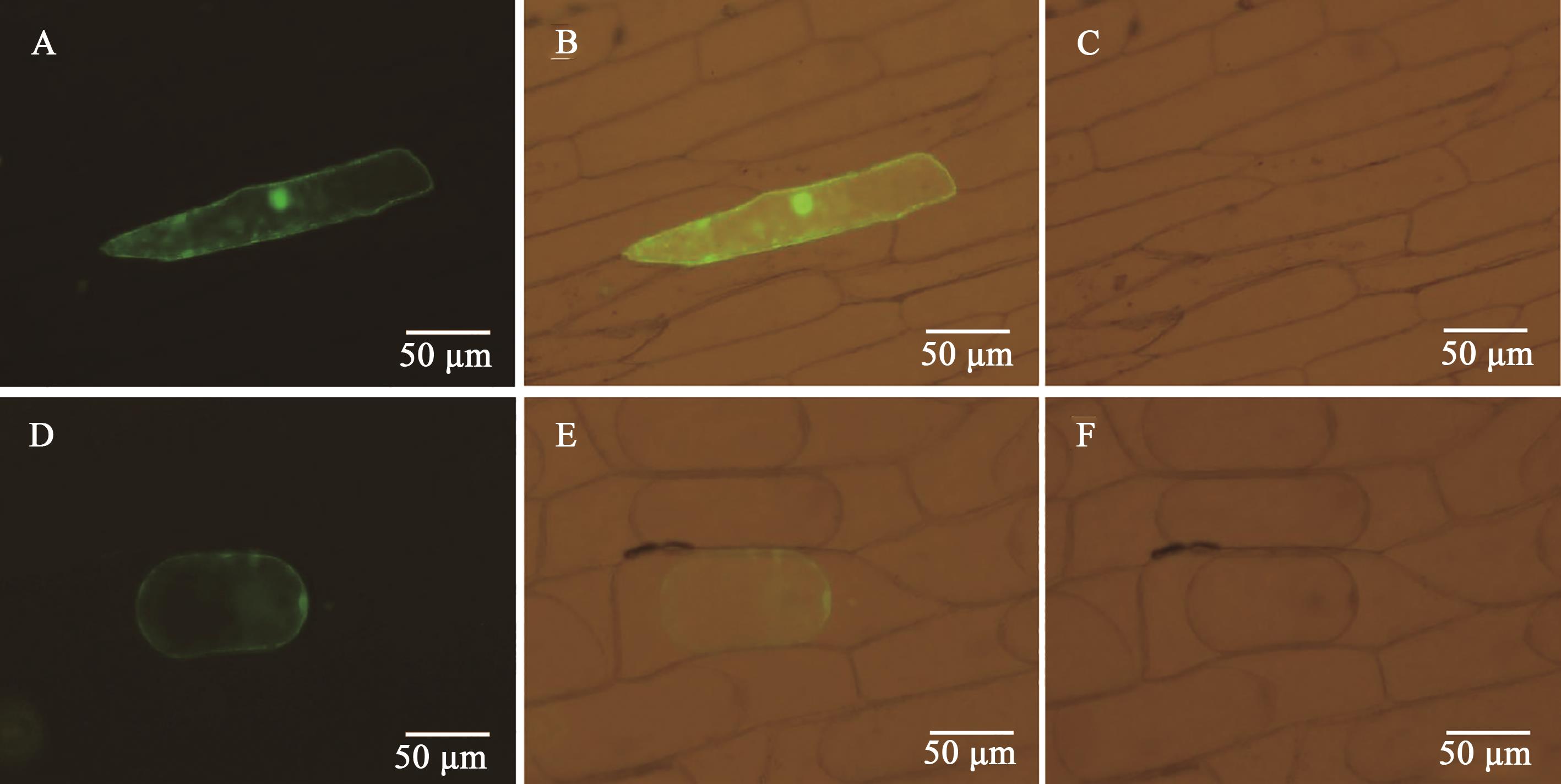
Fig. 3 Subcellular localization of GbGlcAKA, B, C: Cell transfected with pCam::GFP under Blu-ray, visible light superimposed Blu-ray, and visible light; D, E, F: Cell transfected with pCam::GbGlcAK-GFP under Blu-ray, visible light superimposed Blu-ray, and visible light after plasma-wall separation
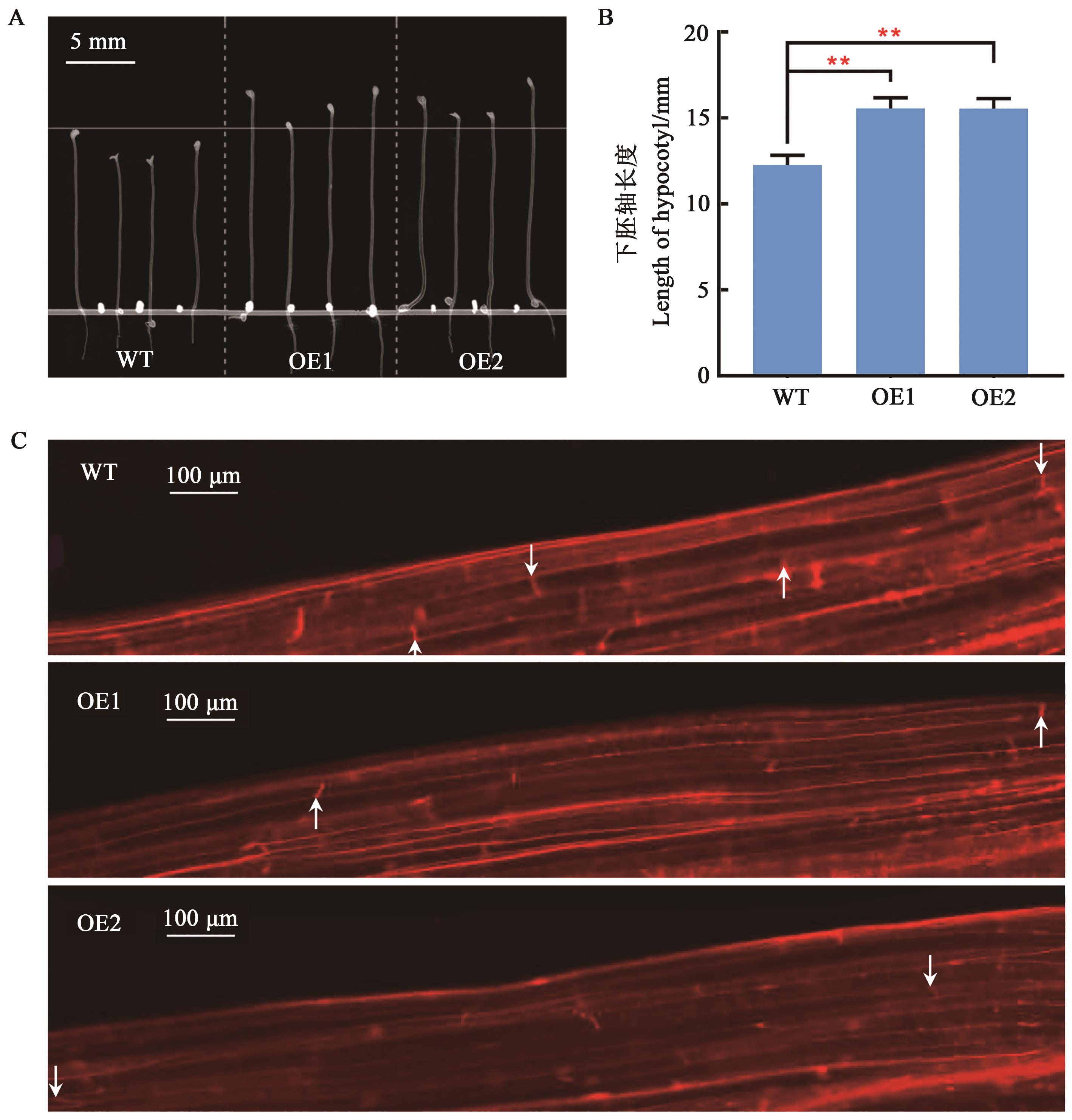
Fig. 5 Analysis of hypocotyl length in transgenic A. thalianaA, B: Comparison of hypocotyl length between transgenic and wild-type Arabidopsis; C: Microscopic observation of hypocotyl cells; ** indicates significant difference at the P<0.01 level (2-tailed t-test);white arrows show the ends of cells
| 1 | 喻树迅,魏晓文,赵新华.中国棉花生产与科技发展 [J].棉花学报, 2000, 12(6): 327-329. |
| YU S X, WEI X W, ZHAO X H. Cotton production and technical development in China [J]. Acta Gossypii Sin., 2000, 12(6): 327-329. | |
| 2 | MA Z, ZHANG Y, WU L, et al.. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement [J]. Nat. Genet., 2021, 53(9): 1385-1391. |
| 3 | BASRA A S, MALIK C P. Development of the cotton fiber [J]. Int. Rev. Cytol., 1984, 89(6): 65-113. |
| 4 | HAIGLER C H, BETANCUR L, STIFF M R, et al.. Cotton fiber: a powerful single-cell model for cell wall and cellulose research [J/OL]. Front. Plant Sci., 2012, 3: 104 [2021-11-09]. . |
| 5 | QIN Y M, ZHU Y X. How cotton fibers elongate: a tale of linear cell-growth mode [J]. Curr. Opin. Plant Biol., 2011, 14(1): 106-111. |
| 6 | SINGH B, AVCI U, EICHLER INWOOD S E, et al.. A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles [J]. Plant Physiol., 2009, 150(2): 684-699. |
| 7 | ZABLACKIS E, JING H, MüLLER B, et al.. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves [J]. Plant Physiol., 1995, 107(4): 1129-1138. |
| 8 | KANTER U, USADEL B, GUERINEAU F, et al.. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides [J]. Planta, 2005, 221(2): 243-254. |
| 9 | LOEWUS F A, KELLY S, NEUFELD E F. Metabolism of myo-inositol in plants: conversion to pectin, hemicellulose, D-xylose, and sugar acids [J]. Proc. Natl. Acad. Sci. USA, 1962, 48(3): 421-425. |
| 10 | NEUFELD E F, FEINGOLD D S, HASSID W Z. Enzymic phosphorylation of D-glucuronic acid by extracts from seedlings of Phaseolus aureus [J]. Arch. Biochem. Biophys., 1959, 83(1): 96-100. |
| 11 | LEIBOWITZ M D, DICKINSON D B, LOEWUS F A, et al.. Partial purification and study of pollen glucuronokinase [J]. Arch. Biochem. Biophys., 1977, 179(2): 559-564. |
| 12 | PIESLINGER A M, HOEPFLINGER M C, TENHAKEN R. Nonradioactive enzyme measurement by high-performance liquid chromatography of partially purified sugar-1-kinase (glucuronokinase) from pollen of Lilium longiflorum [J]. Anal. Biochem., 2009, 388(2): 254-259. |
| 13 | PIESLINGER A M, HOEPFLINGER M C, TENHAKEN R. Cloning of Glucuronokinase from Arabidopsis thaliana, the last missing enzyme of the myo-inositol oxygenase pathway to nucleotide sugars [J]. J. Biol. Chem., 2010, 285(5): 2902-2910. |
| 14 | HOLDEN H M, THODEN J B, TIMSON D J, et al.. Galactokinase: structure, function and role in type Ⅱ galactosemia [J]. Cell. Mol. Life Sci., 2004, 61(19-20): 2471-2484. |
| 15 | XIAO W, HU S, ZHOU X, et al.. A glucuronokinase gene in Arabidopsis, AtGlcAK, is involved in drought tolerance by modulating sugar metabolism [J]. Plant Mol. Biol. Rep., 2017, 35(2): 1-14. |
| 16 | IVANOV KAVKOVA E, BLöCHL C, TENHAKEN R. The Myo-inositol pathway does not contribute to ascorbic acid synthesis [J]. Plant Biol., 2019, 21 (): 95-102. |
| 17 | LIU Z, WANG X, SUN Z, et al.. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes [J/OL]. BMC Plant Biol., 2021, 21(1): 89[2022-03-07]. . |
| 18 | WANG Z, YANG Z, LI F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton [J]. Plant Biotechnol. J., 2019, 17(9): 1706-1722. |
| 19 | 吴立柱,王省芬,李喜焕,等.通用型植物表达载体pCamE的构建及功能验证[J].农业生物技术学报, 2014, 22(6): 661-671. |
| WU L Z, WANG X F, LI X H, et al.. Construction and function identification of universal plant expression vector pCamE [J]. J. Agric. Biotechnol., 2014, 22(6): 661-671. | |
| 20 | WANG M, TU L, YUAN D, et al.. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense [J]. Nat. Genet., 2019, 51(2): 224-229. |
| 21 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al.. Pfam: The protein families database in 2021 [J]. Nucleic Acids Res., 2021, 49(D1): D412-D419. |
| 22 | ARTIMO P, JONNALAGEDDA M, ARNOLD K, et al.. ExPASy: SIB bioinformatics resource portal [J]. Nucleic Acids Res., 2012, 40(W1): W597-W603. |
| 23 | CUTHBERTSON J M, DOYLE D A, SANSOM M S. Transmembrane helix prediction: a comparative evaluation and analysis [J]. Protein Eng. Des. Sel., 2005, 18(6): 295-308. |
| 24 | KROGH A, LARSSON B, GVON HEIJNE, et al.. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes [J]. J. Mol. Biol., 2001, 305(3): 567-580. |
| 25 | MÖLLER S, CRONING M D, APWEILER R. Evaluation of methods for the prediction of membrane spanning regions [J]. Bioinformatics, 2001, 17(7): 646-653. |
| 26 | ALMAGRO ARMENTEROS J J, TSIRIGOS K D, SøNDERBY C K, et al.. SignalP 5.0 improves signal peptide predictions using deep neural networks [J]. Nat. Biotechnol., 2019, 37(4): 420-423. |
| 27 | KUMAR S, STECHER G, TAMURA K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets [J]. Mol. Biol. Evol., 2016, 33(7): 1870-1874. |
| 28 | YANG J, JI L, WANG X, et al.. Overexpression of 3-deoxy-7-phosphoheptulonate synthase gene from Gossypium hirsutum enhances Arabidopsis resistance to Verticillium wilt [J]. Plant Cell Rep., 2015, 34(8): 1429-1441. |
| 29 | CHEN H, NELSON R S, SHERWOOD J L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection [J]. Biotechniques, 1994, 16(4): 664-668, 670. |
| 30 | CLOUGH S J, BENT A F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J., 1998, 16(6): 735-743. |
| 31 | An C, Wang C, Mou Z. The Arabidopsis Elongator complex is required for nonhost resistance against the bacterial pathogens Xanthomonas citri subsp. citri and Pseudomonas syringae pv. phaseolicola NPS3121 [J]. New Phytol., 2017, 214(3): 1245-1259. |
| 32 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4): 402-408. |
| 33 | JEAN-MARC L, TRUNG-BIEU N, BRIGITTE C, et al.. QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations [J]. BMC Plant Biol., 2005, 45(1): 123-140. |
| 34 | SERNA L, MARTIN C. Trichomes: different regulatory networks lead to convergent structures [J]. Trends Plant Sci., 2006, 11(6): 274-280. |
| 35 | GUAN X, YU N, SHANGGUAN X, et al.. Arabidopsis trichome research sheds light on cotton fiber development mechanisms [J]. Chin Sci. Bull., 2007, 52(13): 1734-1741. |
| 36 | GUAN X, PANG M, NAH G, et al.. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development [J/OL]. Nat. Commun., 2014, 5: 3050[2022-03-07]. . |
| 37 | MA Z, HE S, WANG X, et al.. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield [J]. Nat. Genet., 2018, 50(6): 803-813. |
| 38 | GENDREAU E, TRAAS J, DESNOS T, et al.. Cellular basis of hypocotyl growth in Arabidopsis thaliana [J]. Plant Physiol., 1997, 114(1): 295-305. |
| 39 | BORON A K, VISSENBERG K. The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion [J]. Plant Cell Rep., 2014, 33(5): 697-706. |
| 40 | 武耀廷,刘进元.棉纤维细胞发育过程中非纤维素多糖的生物合成 [J].棉花学报, 2004, 16(3): 189-192. |
| WU Y, LIU J. Noncellulosic polysaccharides biosynthesis in cotton fiber developing [J]. Cotton Sci., 2004, 16(3): 189-193. | |
| 41 | 喻树迅,朱玉贤,陈晓亚.棉花纤维发育生物学 [M].北京:科学出版社, 2016: 169. |
| 42 | KROH M, MIKI-HIROSIGE H, ROSEN W, et al.. Incorporation of label into pollen tube walls from myoinositol-labeled Lilium longiflorum pistils [J]. Plant Physiol., 1970, 45(1): 92-94. |
| 43 | SEITZ B, KLOS C, WURM M, et al.. Matrix polysaccharide precursors in Arabidopsis cell walls are synthesized by alternate pathways with organ-specific expression patterns [J]. Plant J., 2000, 21(6): 537-546. |
| [1] | Hao JIA, Hongzhe WANG, Zhengwen SUN, Qishen GU, Dongmei ZHANG, Xingyi WANG, Yan ZHANG, Huaiyu LU, Zhiying MA, Xingfen WANG. Genome-wide Identification of VOZ Genes Family in Cotton and Study on Salt Tolerance Function of GhVOZ1 [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 58-68. |
| [2] | Guiyuan ZHAO, Yongqiang WANG, Jianguang LIU, Zhao GENG, Hanshuang ZHANG, Liqiang WU, Xingfen WANG, Guiyin ZHANG. Effect of Exogenous Gene Insertion Site on Bt Protein Content in Insect-resistant Cotton [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 44-53. |
| [3] | Yixin CHEN, Xiubo YANG, Shijun TIAN, Cong WANG, Zhiying BAI, Cundong LI, Ke ZHANG. Response of GhCOMT28 to Drought Stress in Gossypium hirsutum [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 45-56. |
| [4] | Zhiduo DONG, Qiuping FU, Jian HUANG, Tong QI, Yanbo FU, Kuerban Kaisaier. Analysis of Salt Tolerance Capacity of Xinjiang Cotton Guring Germination [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 57-67. |
| [5] | Zicheng PENG, Hongli DU, Ming WANG, Fenghua ZHANG, Haichang YANG. Research on AMF Regulation of Cotton Growth and Ion Balance Under Salt Alkali Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(2): 33-41. |
| [6] | Songjiang DUAN, Haoran HU, Chengjie ZHANG, Wei SUN, Yifan WU, Rensong GUO, Jusong ZHANG. Differences in Nitrogen Efficiency of Different Genotypes of Island Cotton and Their Effects on Photosynthetic Characteristics and Yield [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 61-71. |
| [7] | Huiting WENG, Haiyang LIU, Huiming GUO, Hongmei CHENG, Jun LI, Chao ZHANG, Xiaofeng SU. Preliminary Function Analysis of GhERF020 Gene in Response to Verticillium Wilt in Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 112-121. |
| [8] | Ziqin LI, Jiaqiang WANG, Zhen LI, Deqiu ZOU, Xiaogong ZHANG, Xiaoyu LUO, Weiyang LIU. Estimation of Chlorophyll Density of Cotton Leaves Based on Spectral Index [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 103-111. |
| [9] | Yukun QIN, Junying CHEN, Lijuan ZHANG. Response of Dry Matter Accumulation Characteristics and Yield of Cotton in North Jiangxi Cotton Region to Nitrogen Reduction Measures [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 191-199. |
| [10] | Ling LIN, Yujie ZHU, Lei FENG, Guangmu TANG, Yunshu ZHANG, Wanli XU. Effects of Aged Cotton Straw Biochars on Soil Properties and Nitrogen Utilization of Wheat [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 184-191. |
| [11] | Jiangbo LI, Wenju GAO, Xiaodong YUN, Jieyin ZHAO, Shiwei GENG, Chunbin HAN, Quanjia CHEN, Qin CHEN. Effects of Different Water Stress Treatments on Core Germplasm Resources of Upland Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 26-39. |
| [12] | Lihua LI, Zhengwen SUN, Huifeng KE, Qishen GU, Liqiang WU, Yan ZHANG, Guiyin ZHANG, Xingfen WANG. Development and Effect Evaluation of KASP Markers for Fiber Strength in Gossypium hirsutum L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 46-55. |
| [13] | Menghua ZHAI, Minghui SUN, Xuerui LI, Xinlong XU, Haizhou GAO, Jusong ZHANG. Effects of DPC on Plant Type Shaping of Cotton Under Different Plant Spacing Configurations [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 145-156. |
| [14] | Zhen CHENG, Jianlong NIU, Yuting MA, weiyang LIU, Xuewei JIANG, Xueqi LIANG, Hongqiang DONG. Dynamic Changes of Cotton Phenological Stages in Alar Reclamation Area of Southern Xinjiang from 1990 to 2020 [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 206-214. |
| [15] | Deyou ZHENG, Dongyun ZUO, Qiaolian WANG, Limin LYU, Hailiang CHENG, Aixing GU, Guoli SONG. Screening of Combination of Flumetralin and Fungicide to Control Cotton Fusarium wilt [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 119-124. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号