




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (1): 38-45.DOI: 10.13304/j.nykjdb.2020.1054
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Qinqin WANG1,2( ), Xiugui CHEN2, Xuke LU2, Shuai WANG2, Yuexin ZHANG2, Yapeng FAN2, Quanjia CHEN1, Wuwei YE1,2(
), Xiugui CHEN2, Xuke LU2, Shuai WANG2, Yuexin ZHANG2, Yapeng FAN2, Quanjia CHEN1, Wuwei YE1,2( )
)
Received:2020-12-11
Accepted:2021-03-03
Online:2022-01-15
Published:2022-01-25
Contact:
Wuwei YE
王琴琴1,2( ), 陈修贵2, 陆许可2, 王帅2, 张悦新2, 范亚朋2, 陈全家1, 叶武威1,2(
), 陈修贵2, 陆许可2, 王帅2, 张悦新2, 范亚朋2, 陈全家1, 叶武威1,2( )
)
通讯作者:
叶武威
作者简介:王琴琴 E-mail: wqq4024@163.com;
基金资助:CLC Number:
Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton[J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45.
王琴琴, 陈修贵, 陆许可, 王帅, 张悦新, 范亚朋, 陈全家, 叶武威. 陆地棉GhPKE1的生物信息学分析及功能验证[J]. 中国农业科技导报, 2022, 24(1): 38-45.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2020.1054
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| KL-GhPKE1 | F:GAGAACACGGGGGACTCTAGAATGGCTGAAAAGGTGACGATAATGG |
| R:CGATCGGGGAAATTCGAGCTCTTACATGATCGAGCAACCTTGTGGG | |
| V-GhPKE1 | F:AGAAGGCCTCCATGGGGATCCATGGCTGAAAAGGTGACGATAATGG |
| R:GAGACGCGTGAGCTCGGTACCTCTTCTCACCGTCTTTGGGCTTTTC | |
| q-GhPKE1 | F:ACAAGTTATGTTACAAAGGTGGTGG |
| R:TCTCAGCTTCTTTGGGTTTTTCCGG | |
GhHistone3 (AF024716) | F:TCAAGACTGATTTGCGTTTCCA |
| R:GCGCAAAGGTTGGTGTCTTC |
Table 1 Primer sequence of GhPKE1
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| KL-GhPKE1 | F:GAGAACACGGGGGACTCTAGAATGGCTGAAAAGGTGACGATAATGG |
| R:CGATCGGGGAAATTCGAGCTCTTACATGATCGAGCAACCTTGTGGG | |
| V-GhPKE1 | F:AGAAGGCCTCCATGGGGATCCATGGCTGAAAAGGTGACGATAATGG |
| R:GAGACGCGTGAGCTCGGTACCTCTTCTCACCGTCTTTGGGCTTTTC | |
| q-GhPKE1 | F:ACAAGTTATGTTACAAAGGTGGTGG |
| R:TCTCAGCTTCTTTGGGTTTTTCCGG | |
GhHistone3 (AF024716) | F:TCAAGACTGATTTGCGTTTCCA |
| R:GCGCAAAGGTTGGTGTCTTC |
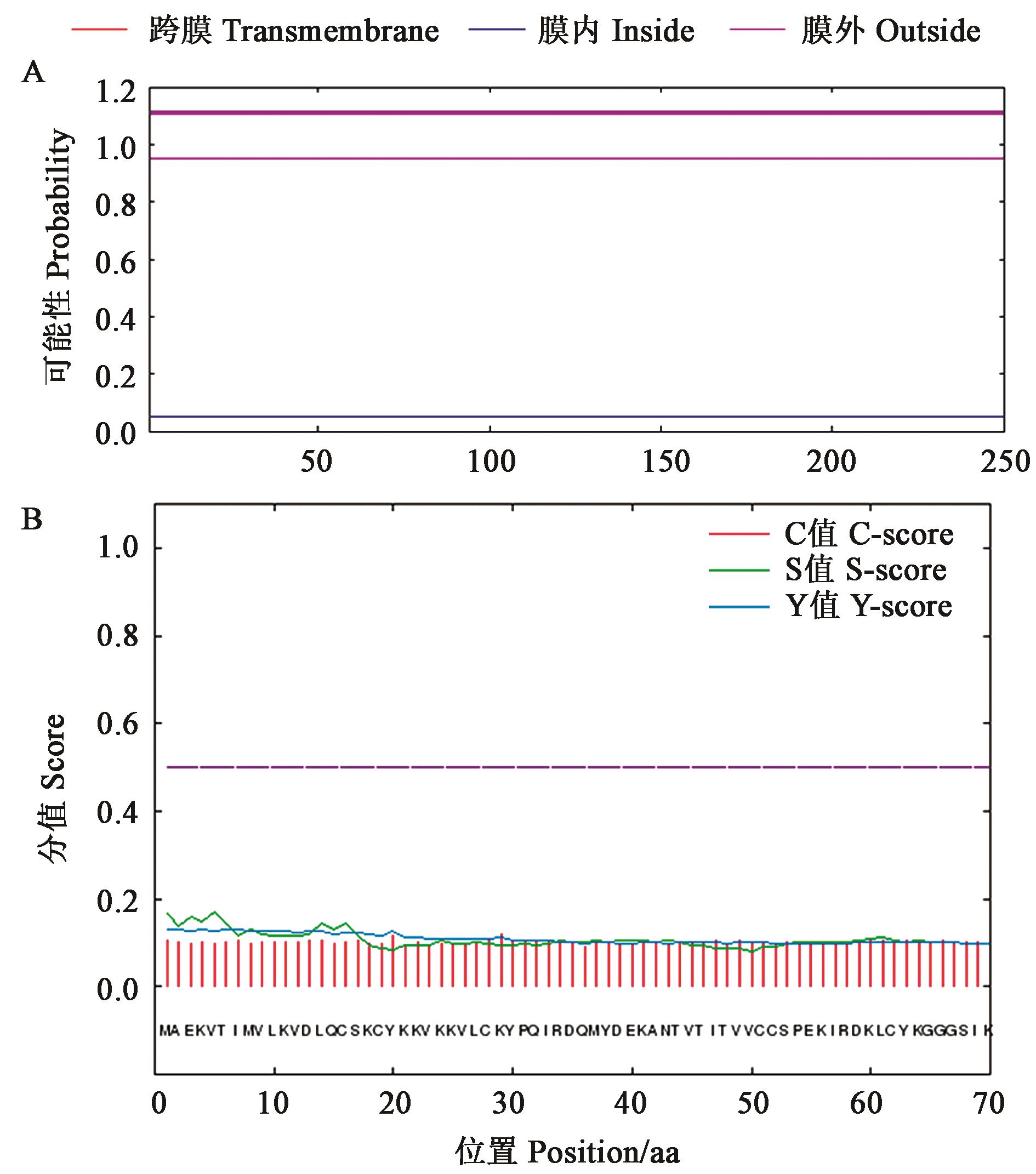
Fig.2 Prediction of transmembrane domain and signal peptide in GhPKE1 proteinA: Transmembrane domain analysis of GhPKE1 protein; B: Signal peptide prediction of GhPKE1 protein

Fig.3 GhPKE1 bioinformatics analysisA: Secondary structure of GhPKE1 protein; B: Prediction of GhPKE1 protein globulin region and disorder region; C: Secondary structure of GhPKE1 protein; D: Interaction network of GhPKE1 protein in Arabidopsis thaliana
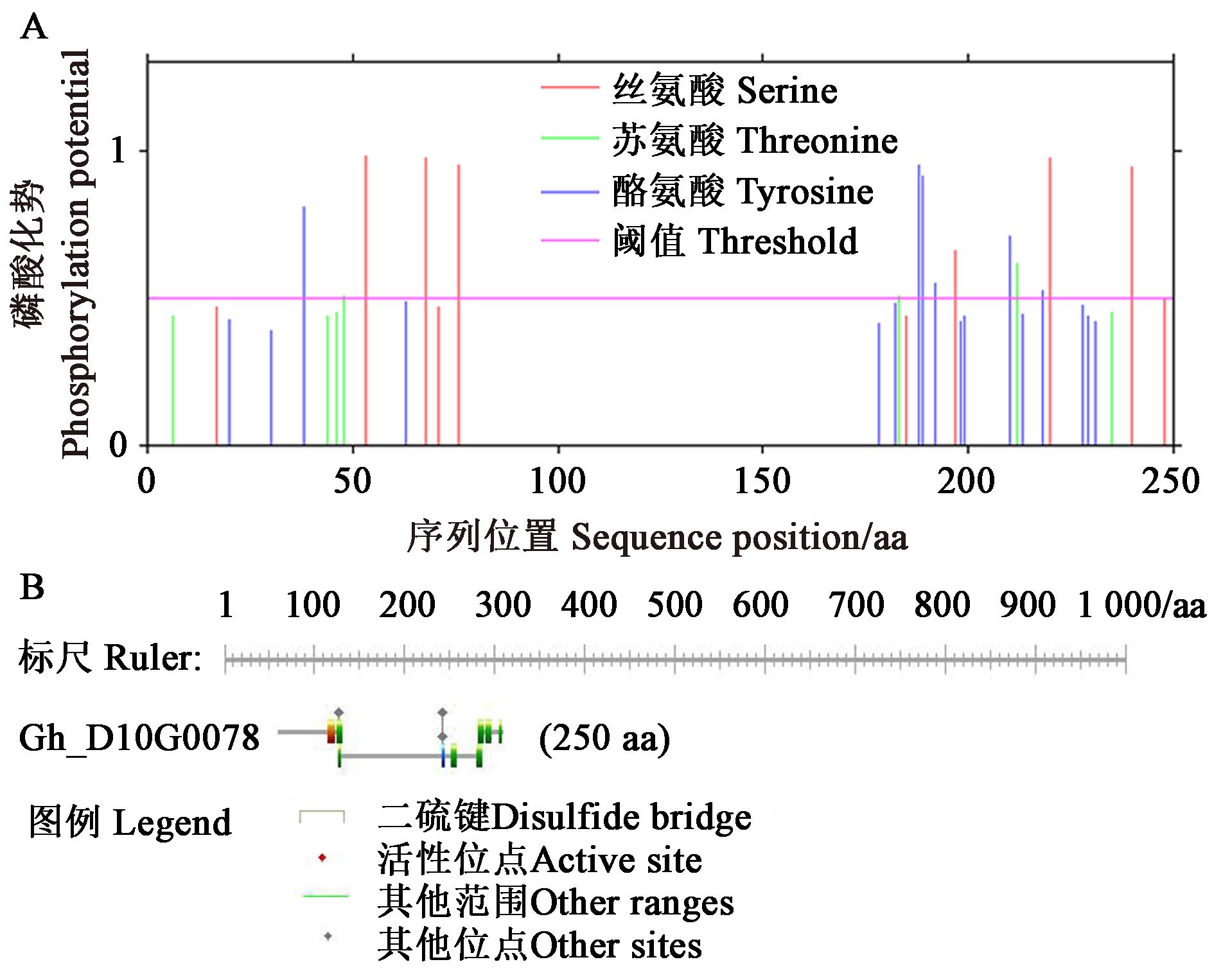
Fig.4 Prediction of GhPKE1 phosphorylation sites and functional areaA: NetPhos 3.1 Server predicts GhPKE1 phosphorylation sites; B: PROSITE predicts GhPKE1 functional area
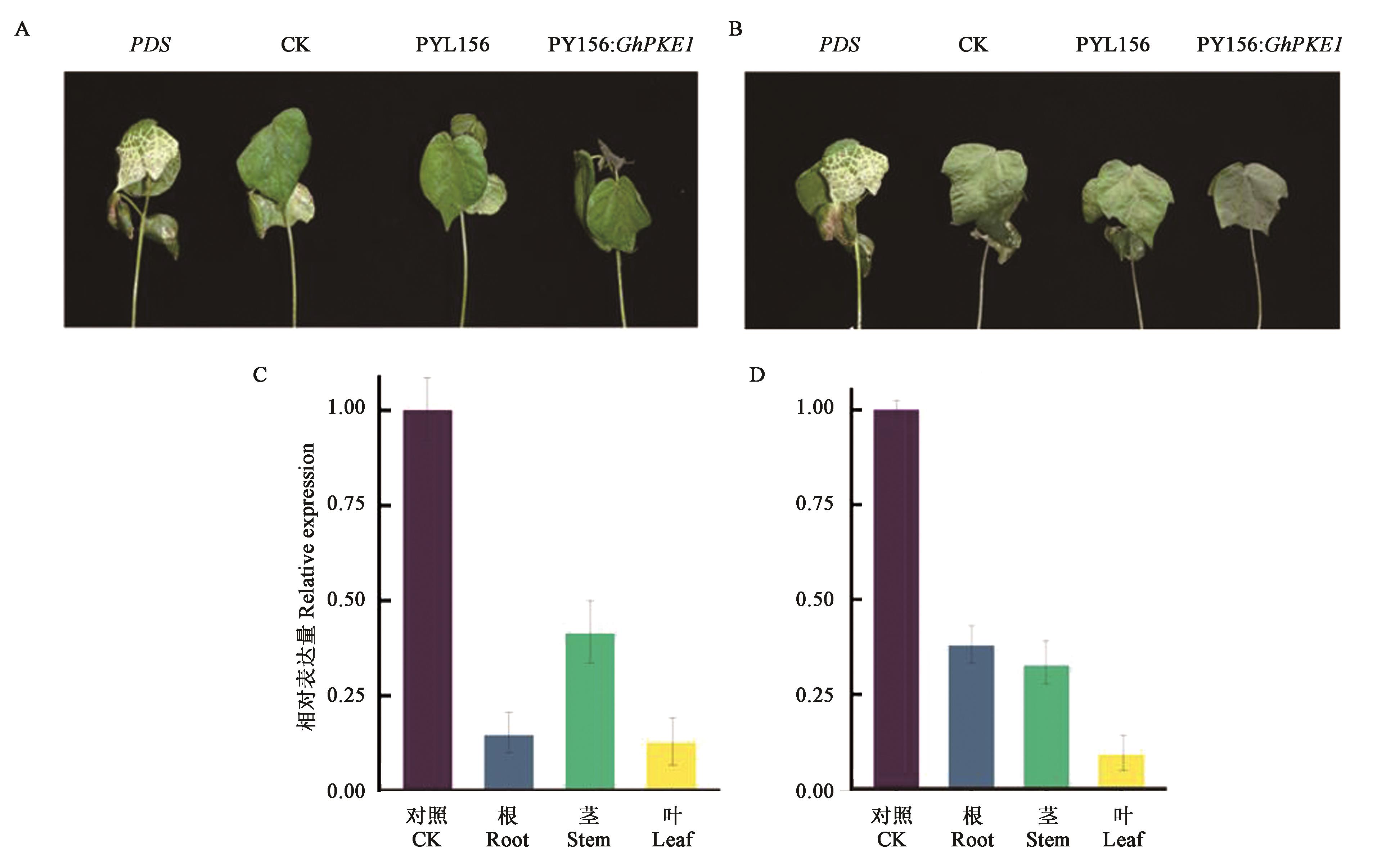
Fig.5 Cotton phenotype and the relative expression level of GhPKE1 after VIGS infectionA: Cotton phenotype under Na2SO4 stress for 12 h; B: Cotton phenotype under CdCl2 stress for 12 h; C: Relative expression level of GhPKE1 after Na2SO4 stress; D: Relative expression level of GhPKE1 after CdCl2 stress
| 1 | 梁丽琛,刘维涛,张雪,等.盐土植物提取修复重金属污染盐土研究进展[J].农业环境科学学报,2016,35(7):1233-1241. |
| LIANG L S, LIU W T, ZHANG X, et al.. Research progress in phytoextraction of heavy metal contaminated saline soil [J]. J. Agro-Environ. Sci., 2016, 35(7):1233-1241. | |
| 2 | 叶武威,穆敏,王俊娟,等.真菌的耐盐基因挖掘与棉花的耐盐性改良研究[C]// 中国农学会棉花分会2016年年会论文汇编.江苏 徐州:中国农业学会,2016:73-74. |
| 3 | KOLEVA-VALKOVA L, VASILEV A. Physiological parameters of young cotton plants, grown on heavy metal contaminated soils [J]. Agrarni Nauki, 2015, 7(18):61-66. |
| 4 | MANOUSAKI E, KALOGERAKIS N. Halophytes-an emerging trend in phytoremediation [J]. Int. J. Phytorem., 2011, 13(10):959-969. |
| 5 | ARIF N, YADAV V, SINGH S, et al.. Influence of high and low levels of plant-beneficial heavy metal ions on plant growth and development [J]. Front Environ. Sci., 2016, 4:69-80. |
| 6 | SALAM L B, OBAYORI O S, ILORI M O, et al.. Effects of cadmium perturbation on the microbial community structure and heavy metal resistome of a tropical agricultural soil [J]. Bioresour. Bioprocess., 2020, 7(1):1-19. |
| 7 | FARCASANU I C, POPA C V, RUTA L L. Calcium and cell response to heavy metals: can yeast provide an answer ? [J]. Calcium Signal Transduction, 2018:23-41. |
| 8 | JALMI S K, BHAGAT P K, VERMA D, et al.. Traversing the links between heavy metal stress and plant signaling [J/OL]. Front Plant Sci., 2018, 9:12 [2020-11-10]. . |
| 9 | EMAMVERDIAN A, DING Y, MOKHBERDORAN F, et al.. Heavy metal stress and some mechanisms of plant defense response [J/OL]. Sci. World J., 2015(2015):756120 [2020-11-10]. . |
| 10 | CHAUDHARY K, AGARWAL S, KHAN S. Role of Phytochelatins (PCs), Metallothioneins (MTs), and Heavy Metal ATPase (HMA) Genes in Heavy Metal Tolerance [M]. Mycoremediation and Environmental Sustainability, Springer, 2018:39-60. |
| 11 | GHORI N, GHORI T, HAYAT M Q, et al.. Heavy metal stress and responses in plants [J]. Int. J. Environ. Sci.Technol., 2019, 16(3):1807-1828. |
| 12 | BELYKH E S, MAYSTRENKO T A, VELEGZHANINOV I O. Recent trends in enhancing the resistance of cultivated plants to heavy metal stress by transgenesis and transcriptional programming [J]. Mol. Biotechnol., 2019, 61(4):725-741. |
| 13 | SHARMA S S, DIETZ K J, MIMURA T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants [J]. Plant Cell Environ., 2016, 39(5):1112-1126. |
| 14 | LI J, CHEN C, WEI J, et al.. SpPKE1, a multiple stress-responsive gene confers salt tolerance in tomato and tobacco [J]. Int. J. Mol. Sci., 2019, 20(10):2478-2493. |
| 15 | PETERSENSUP SUP S K E SJAN. The lysine-rich motif of intrinsically disordered stress protein CDeT11-24 from Craterostigma plantagineum is responsible for phosphatidic acid binding and protection of enzymes from damaging effects caused by desiccation [J]. J. Exp. Bot., 2012, 63(13):4919-4929. |
| 16 | PITZSCHKE A, XUE H, PERSAK H, et al.. Post-translational modification and secretion of azelaic acid induced 1 (AZI1), a hybrid proline-rich protein from Arabidopsis [J/OL]. Int. J. Mol. Sci., 2016, 17(1):85 [2020-11-10]. . |
| 17 | BENEŠ V, LEONHARDT T, SÁCKÝ J, et al.. Two P1B-1-ATPases of Amanita strobiliformis with distinct properties in Cu/Ag transport [J/OL]. Front Microbiol., 2018, 9:747 [2020-11-10]. . |
| 18 | ZHANG Y, CHEN K, ZHAO F, et al.. OsATX1 interacts with heavy metal P1B-type ATPases and affects copper transport and distribution [J]. Plant Physiol., 2018, 178(1):329-344. |
| 19 | DAI W, WANG M, GONG X, et al.. The transcription factor Fc WRKY 40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs [J]. New Phytol., 2018, 219(3):972-989. |
| 20 | CONG W, MIAO Y, XU L, et al.. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.) [J/OL]. BMC Plant Biol., 2019, 19(1):7 [2020-11-10]. . |
| 21 | WANG X, GONG X, CAO F, et al.. HvPAA1 encodes a P-Type ATPase, a novel gene for cadmium accumulation and tolerance in barley (Hordeum vulgare L.) [J/OL]. Int. J. Mol. Sci., 2019, 20(7):1732 [2020-11-10]. . |
| 22 | KEERAN N S, GANESAN G, PARIDA A K. A novel heavy metal ATPase peptide from Prosopis juliflora is involved in metal uptake in yeast and tobacco [J]. Transgenic Res., 2017, 26(2):247-261. |
| 23 | LEKEUX G, CROWET J, NOUET C, et al.. Homology modeling and in vivo functional characterization of the zinc permeation pathway in a heavy metal P-type ATPase [J]. J. Exp. Bot., 2019, 70(1):329-341. |
| 24 | GRISPEN V M, HAKVOORT H W, BLIEK T, et al.. Combined expression of the Arabidopsis metallothionein MT2b and the heavy metal transporting ATPase HMA4 enhances cadmium tolerance and the root to shoot translocation of cadmium and zinc in tobacco [J]. Environ. Exp. Bot., 2011, 72(1):71-76. |
| 25 | HUSSAIN D, HAYDON M J, WANG Y, et al.. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis [J]. Plant Cell, 2004, 16(5):1327-1339. |
| 26 | SHRIVASTAVA M, KHANDELWAL A, SRIVASTAVA S. Heavy Metal Hyperaccumulator Plants: The Resource to Understand the Extreme Adaptations of Plants Towards Heavy Metals [M]. Plant-Metal Interactions, Springer, 2019:79-97. |
| 27 | LI J, WANG Y, WEI J, et al.. A tomato proline-, lysine-, and glutamic-rich type gene SpPKE1 positively regulates drought stress tolerance [J]. Biochem. Biophys. Res. Commun., 2018, 499(4):777-782. |
| [1] | Yanqin MA, Yujie ZHOU, Haicheng LONG, Ju LI, Haie WANG, Wei CHANG, Zhi LI, Jian ZHONG, Mingjun MIAO, Liang YANG. Construction of TRV-mediated VIGS System in Brassica rapa subsp. chinensis and Brassica juncea [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 239-249. |
| [2] | Zhining YANG, Xuke LU, Yapeng FAN, Yuping SUN, Xin YU, Liang WANG, Yongjian GAO, Gulijiayinashen Habuli, Gulishaxi Nasiyi, Wumuer Abudu, Hui HUANG, Menghao ZHANG, Lidong WANG, Xiao CHEN, Lei XIAO, Xinrui ZHANG, Shuai WANG, Xiugui CHEN, Junjuan WANG, Lixue GUO, Wenwei GAO, Wuwei YE. Response Mechanism of Cotton GhDMT7 Gene to 5-azacytidine [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 28-35. |
| [3] | Jinxian LIU, Lijuan WANG, Jie LIU, Xianyu FU, Guangheng WU. Identification and Expression Analysis of Calmodulin-binding Transcription Activator (CAMTA) Family Genes in Tea Plants [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 71-82. |
| [4] | Huiting WENG, Haiyang LIU, Huiming GUO, Hongmei CHENG, Jun LI, Chao ZHANG, Xiaofeng SU. Preliminary Function Analysis of GhERF020 Gene in Response to Verticillium Wilt in Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 112-121. |
| [5] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [6] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [7] | Bo LIU, Wangtian WANG, Li MA, Junyan WU, Yuanyuan PU, Lijun LIU, Yan FANG, Wancang SUN, Yan ZHANG, Ruimin LIU, Xiucun ZENG. Identification and Characterization of IPT Gene Family in Brassica rapa L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 56-66. |
| [8] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [9] | Man ZHANG, Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG. Cloning and Functional Analysis of GhNAC1 in Upland Cotton Involved in Verticillium Wilt Resistance [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 35-44. |
| [10] | Mengyuan HAO, Qi HANG, Gongyao SHI. Application and Prospect of Virus-induced Gene Silencing in Crop Gene Function Research [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 1-13. |
| [11] | GUAN Sijing, WANG Nan, XU Rongrong, GE Tiantian, GAO Jing, YAN Yonggang, ZHANG Gang, CHEN Ying, ZHANG Mingying. [J]. Journal of Agricultural Science and Technology, 2021, 23(12): 66-75. |
| [12] | XU Mengjun, GAO Tian, WANG Pengfei, LI Gezi, KANG Guozhang*. Function of Calcium Dependent Protein Kinase 34 in Grain Starch Synthesis of Wheat (Triticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(2): 26-33. |
| [13] | LI Xia1,2, FAN Xin2, LIANG Chengzhen2, WANG Yuan2, ZHANG Rui2, MENG Zhigang2*, LIU Xiaodong1*, SUN Guoqing2*. Functional Analysis of BS2 Gene in Tobacco [J]. Journal of Agricultural Science and Technology, 2019, 21(11): 43-50. |
| [14] | YANG Xiaomin, RUI Cun, ZHANG Yuexin, WANG Delong, WANG Junjuan, LU Xuke, CHEN Xiugui, GUO Lixue, WANG Shuai, CHEN Chao, YE Wuwei*. Cloning and Stress Resistance Analysis of Cotton DNA Methyltransferase GhDMT9 Gene [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 12-19. |
| [15] | ZHAO Yan1, SHENG Yunlong1, SONG Yafei1, ZHANG Jiakuo1, WENG Qiaoyun1, YUAN Jincheng1, ZHAO Zhihai2*, LIU Yinghui1*. Genome-wide Identification and Bio-informatics Analysis of Superoxide Dismutase Gene Family in Setaria italica [J]. Journal of Agricultural Science and Technology, 2018, 20(8): 1-6. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号