




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (4): 67-76.DOI: 10.13304/j.nykjdb.2021.0794
收稿日期:2021-09-08
接受日期:2021-10-20
出版日期:2023-04-01
发布日期:2023-06-26
通讯作者:
田洁
作者简介:闫艺薇 E-mail: yanyw0102@163.com;
基金资助:Received:2021-09-08
Accepted:2021-10-20
Online:2023-04-01
Published:2023-06-26
Contact:
Jie TIAN
摘要:
为挖掘大蒜NAC转录因子家族成员,研究其在低温胁迫下的表达特性,基于大蒜转录组的测序结果,对低温胁迫下大蒜NAC转录因子家族的响应进行分析。结果表明,共鉴定出49个大蒜NAC基因,根据系统发育特征将其分为10个亚族。大蒜NAC蛋白的序列长度为81~619 aa,分子量9 293.66~70 229.04 Da,且均为亲水蛋白。保守结构域分析发现,大部分NAC蛋白在N端具有保守性,其中18个NAC蛋白在N端均有motif 3、motif 4、motif 1、motif 2、motif 5保守域。从中随机挑选6个蛋白(AsNAC001、AsNAC010、AsNAC013、AsNAC014、AsNAC017、AsNAC047)对其进行蛋白二级及三级结构的预测,发现无规则卷曲是构成大蒜NAC蛋白的重要组成部分,其次是α-螺旋和延长链,且这6个NAC蛋白具有高度的相似性。实时荧光定量分析发现,检测的6个NAC基因中有4个(AsNAC001、AsNAC010、AsNAC013、AsNAC014)在低温胁迫12和24 h时显著上调表达;2个(AsNAC017、AsNAC047)在低温胁迫4 h时显著下调表达,其中AsNAC010基因的表达量显著高于其他基因,说明NAC转录因子在大蒜响应低温胁迫的过程中起重要调控作用,为大蒜NAC蛋白的生理调控功能的分析奠定了理论基础。
中图分类号:
闫艺薇, 田洁. 大蒜NAC基因家族的鉴定与低温表达分析[J]. 中国农业科技导报, 2023, 25(4): 67-76.
Yiwei YAN, Jie TIAN. Identification and Expression Analysis of NAC Gene Family Under Low Temperature in Allium sativum L.[J]. Journal of Agricultural Science and Technology, 2023, 25(4): 67-76.
基因名称 Gene name | 引物序列 Primer sequence(5’-3’) | 退火温度 Annealing temperature/℃ |
|---|---|---|
| AsNAC001 | F: CCGAGAAGGCATTATTTG | 52 |
| R: ACCCGTCGCTTTCCAGTA | ||
| AsNAC010 | F: TCAACCTCTACAAGCACGACC | 56 |
| R: GAACCCTTTGGGCACGAT | ||
| AsNAC013 | F: CTTCTACAGCCTTCGTG | 50 |
| R: AGTCTTCCTCATCCCTAC | ||
| AsNAC014 | F: ATCTTGTGATGCCTGGTT | 49 |
| R: AATGGCTGCTAATGCTG | ||
| AsNAC017 | F: TCCAGCAGGGTTCAGATT | 54 |
| R: ACTCCCAAGGCTCACAAG | ||
| AsNAC047 | F: AAAATCCTCATACACCGCCTAA | 57 |
| R: TCGTCCCGAAGCATCCAC | ||
| CYP | F: AAGGACGAGAACTTCATC | — |
| R: TCAATATCTCTCACCACTTC |
表1 AsNAC 基因实时荧光定量PCR引物序列
Table 1 Primer sequences of AsNAC genes for real-time fluorescence quantification
基因名称 Gene name | 引物序列 Primer sequence(5’-3’) | 退火温度 Annealing temperature/℃ |
|---|---|---|
| AsNAC001 | F: CCGAGAAGGCATTATTTG | 52 |
| R: ACCCGTCGCTTTCCAGTA | ||
| AsNAC010 | F: TCAACCTCTACAAGCACGACC | 56 |
| R: GAACCCTTTGGGCACGAT | ||
| AsNAC013 | F: CTTCTACAGCCTTCGTG | 50 |
| R: AGTCTTCCTCATCCCTAC | ||
| AsNAC014 | F: ATCTTGTGATGCCTGGTT | 49 |
| R: AATGGCTGCTAATGCTG | ||
| AsNAC017 | F: TCCAGCAGGGTTCAGATT | 54 |
| R: ACTCCCAAGGCTCACAAG | ||
| AsNAC047 | F: AAAATCCTCATACACCGCCTAA | 57 |
| R: TCGTCCCGAAGCATCCAC | ||
| CYP | F: AAGGACGAGAACTTCATC | — |
| R: TCAATATCTCTCACCACTTC |
基因名称 Gene name | 氨基酸长度 Number of amino acid/aa | 编码区长度 Length of cDNA/bp | 等电点 Theoretical pI | 分子量 Molecular weight/Da | 不稳定指数 Instability index | 脂溶系数 Aliphatic index | 总平均疏水指数Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| AsNAC001 | 317 | 966 | 5.48 | 36 739.14 | 40.50 | 60.60 | -0.939 |
| AsNAC002 | 227 | 692 | 9.18 | 25 415.27 | 48.38 | 56.65 | -0.654 |
| AsNAC003 | 314 | 966 | 6.22 | 36 414.20 | 35.74 | 67.36 | -0.662 |
| AsNAC004 | 299 | 911 | 6.34 | 34 235.70 | 29.90 | 57.06 | -0.716 |
| AsNAC005 | 297 | 905 | 4.98 | 34 391.81 | 44.56 | 64.31 | -0.904 |
| AsNAC006 | 181 | 555 | 9.86 | 20 697.99 | 25.75 | 77.07 | -0.480 |
| AsNAC007 | 284 | 866 | 5.93 | 32 876.85 | 38.61 | 63.84 | -0.816 |
| AsNAC008 | 174 | 533 | 9.83 | 19 776.87 | 33.91 | 74.08 | -0.466 |
| AsNAC009 | 295 | 899 | 8.61 | 33 523.98 | 39.65 | 65.15 | -0.637 |
| AsNAC010 | 220 | 670 | 7.17 | 25 613.94 | 35.54 | 66.45 | -0.780 |
| AsNAC011 | 81 | 247 | 9.79 | 9 293.66 | 29.75 | 53.09 | -0.793 |
| AsNAC012 | 350 | 1 067 | 4.95 | 40 488.80 | 43.51 | 69.63 | -0.827 |
| AsNAC013 | 299 | 914 | 8.07 | 34 120.71 | 37.43 | 63.24 | -0.581 |
| AsNAC014 | 311 | 948 | 6.83 | 35 302.75 | 45.96 | 74.63 | -0.591 |
| AsNAC015 | 153 | 466 | 9.60 | 17 763.48 | 36.73 | 72.61 | -0.460 |
| AsNAC016 | 142 | 433 | 9.54 | 16 467.63 | 30.22 | 68.59 | -0.706 |
| AsNAC017 | 241 | 735 | 6.17 | 27 870.55 | 40.27 | 59.83 | -0.651 |
| AsNAC018 | 234 | 713 | 5.74 | 27 139.67 | 41.73 | 55.77 | -0.631 |
| AsNAC019 | 196 | 597 | 8.62 | 22 554.44 | 21.80 | 61.68 | -0.671 |
| AsNAC020 | 256 | 780 | 6.34 | 28 965.66 | 40.18 | 63.52 | -0.550 |
| AsNAC021 | 205 | 625 | 8.75 | 23 361.40 | 38.21 | 60.78 | -0.573 |
| AsNAC022 | 185 | 567 | 9.45 | 21 445.02 | 20.73 | 72.16 | -0.374 |
| AsNAC023 | 159 | 484 | 9.47 | 18 236.02 | 23.81 | 61.95 | -0.664 |
| AsNAC024 | 151 | 460 | 9.40 | 17 500.04 | 36.29 | 57.48 | -0.788 |
| AsNAC025 | 147 | 448 | 9.70 | 16 631.15 | 31.41 | 69.12 | -0.589 |
| AsNAC026 | 322 | 985 | 8.94 | 36 669.70 | 40.31 | 68.17 | -0.567 |
| AsNAC027 | 256 | 780 | 8.57 | 29 357.41 | 30.19 | 61.37 | -0.656 |
| AsNAC028 | 144 | 439 | 9.10 | 16 917.46 | 34.03 | 56.25 | -0.816 |
| AsNAC029 | 156 | 475 | 9.67 | 17 769.38 | 43.29 | 73.78 | -0.574 |
| AsNAC030 | 198 | 603 | 5.25 | 22 305.54 | 44.75 | 62.47 | -0.819 |
| AsNAC031 | 619 | 1 887 | 4.78 | 70 229.04 | 46.44 | 70.39 | -0.709 |
| AsNAC032 | 176 | 542 | 9.19 | 20 419.75 | 38.67 | 73.64 | -0.274 |
| AsNAC033 | 238 | 728 | 9.17 | 27 182.77 | 34.76 | 60.21 | -0.607 |
| AsNAC034 | 272 | 835 | 7.80 | 31 081.00 | 43.57 | 64.89 | -0.604 |
| AsNAC035 | 209 | 637 | 7.00 | 23 813.71 | 30.07 | 73.68 | -0.678 |
| AsNAC036 | 276 | 841 | 4.95 | 31 051.17 | 32.52 | 67.50 | -0.791 |
表2 大蒜NAC蛋白的理化性质
Table 2 Physical and chemical properties of NAC proteins in Allium sativum L.
基因名称 Gene name | 氨基酸长度 Number of amino acid/aa | 编码区长度 Length of cDNA/bp | 等电点 Theoretical pI | 分子量 Molecular weight/Da | 不稳定指数 Instability index | 脂溶系数 Aliphatic index | 总平均疏水指数Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| AsNAC001 | 317 | 966 | 5.48 | 36 739.14 | 40.50 | 60.60 | -0.939 |
| AsNAC002 | 227 | 692 | 9.18 | 25 415.27 | 48.38 | 56.65 | -0.654 |
| AsNAC003 | 314 | 966 | 6.22 | 36 414.20 | 35.74 | 67.36 | -0.662 |
| AsNAC004 | 299 | 911 | 6.34 | 34 235.70 | 29.90 | 57.06 | -0.716 |
| AsNAC005 | 297 | 905 | 4.98 | 34 391.81 | 44.56 | 64.31 | -0.904 |
| AsNAC006 | 181 | 555 | 9.86 | 20 697.99 | 25.75 | 77.07 | -0.480 |
| AsNAC007 | 284 | 866 | 5.93 | 32 876.85 | 38.61 | 63.84 | -0.816 |
| AsNAC008 | 174 | 533 | 9.83 | 19 776.87 | 33.91 | 74.08 | -0.466 |
| AsNAC009 | 295 | 899 | 8.61 | 33 523.98 | 39.65 | 65.15 | -0.637 |
| AsNAC010 | 220 | 670 | 7.17 | 25 613.94 | 35.54 | 66.45 | -0.780 |
| AsNAC011 | 81 | 247 | 9.79 | 9 293.66 | 29.75 | 53.09 | -0.793 |
| AsNAC012 | 350 | 1 067 | 4.95 | 40 488.80 | 43.51 | 69.63 | -0.827 |
| AsNAC013 | 299 | 914 | 8.07 | 34 120.71 | 37.43 | 63.24 | -0.581 |
| AsNAC014 | 311 | 948 | 6.83 | 35 302.75 | 45.96 | 74.63 | -0.591 |
| AsNAC015 | 153 | 466 | 9.60 | 17 763.48 | 36.73 | 72.61 | -0.460 |
| AsNAC016 | 142 | 433 | 9.54 | 16 467.63 | 30.22 | 68.59 | -0.706 |
| AsNAC017 | 241 | 735 | 6.17 | 27 870.55 | 40.27 | 59.83 | -0.651 |
| AsNAC018 | 234 | 713 | 5.74 | 27 139.67 | 41.73 | 55.77 | -0.631 |
| AsNAC019 | 196 | 597 | 8.62 | 22 554.44 | 21.80 | 61.68 | -0.671 |
| AsNAC020 | 256 | 780 | 6.34 | 28 965.66 | 40.18 | 63.52 | -0.550 |
| AsNAC021 | 205 | 625 | 8.75 | 23 361.40 | 38.21 | 60.78 | -0.573 |
| AsNAC022 | 185 | 567 | 9.45 | 21 445.02 | 20.73 | 72.16 | -0.374 |
| AsNAC023 | 159 | 484 | 9.47 | 18 236.02 | 23.81 | 61.95 | -0.664 |
| AsNAC024 | 151 | 460 | 9.40 | 17 500.04 | 36.29 | 57.48 | -0.788 |
| AsNAC025 | 147 | 448 | 9.70 | 16 631.15 | 31.41 | 69.12 | -0.589 |
| AsNAC026 | 322 | 985 | 8.94 | 36 669.70 | 40.31 | 68.17 | -0.567 |
| AsNAC027 | 256 | 780 | 8.57 | 29 357.41 | 30.19 | 61.37 | -0.656 |
| AsNAC028 | 144 | 439 | 9.10 | 16 917.46 | 34.03 | 56.25 | -0.816 |
| AsNAC029 | 156 | 475 | 9.67 | 17 769.38 | 43.29 | 73.78 | -0.574 |
| AsNAC030 | 198 | 603 | 5.25 | 22 305.54 | 44.75 | 62.47 | -0.819 |
| AsNAC031 | 619 | 1 887 | 4.78 | 70 229.04 | 46.44 | 70.39 | -0.709 |
| AsNAC032 | 176 | 542 | 9.19 | 20 419.75 | 38.67 | 73.64 | -0.274 |
| AsNAC033 | 238 | 728 | 9.17 | 27 182.77 | 34.76 | 60.21 | -0.607 |
| AsNAC034 | 272 | 835 | 7.80 | 31 081.00 | 43.57 | 64.89 | -0.604 |
| AsNAC035 | 209 | 637 | 7.00 | 23 813.71 | 30.07 | 73.68 | -0.678 |
| AsNAC036 | 276 | 841 | 4.95 | 31 051.17 | 32.52 | 67.50 | -0.791 |
基因名称 Gene name | 氨基酸长度 Number of amino acid/aa | 编码区长度 Length of cDNA/bp | 等电点 Theoretical pI | 分子量 Molecular weight/Da | 不稳定指数 Instability index | 脂溶系数 Aliphatic index | 总平均疏水指数Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| AsNAC037 | 290 | 884 | 6.45 | 33 210.39 | 45.73 | 63.59 | -0.694 |
| AsNAC038 | 176 | 548 | 9.74 | 20 481.63 | 39.16 | 79.77 | -0.354 |
| AsNAC039 | 147 | 448 | 9.72 | 16 705.32 | 31.72 | 70.41 | -0.582 |
| AsNAC040 | 179 | 555 | 9.74 | 21 143.43 | 51.78 | 68.60 | -0.450 |
| AsNAC041 | 152 | 463 | 7.78 | 17 151.33 | 42.10 | 56.51 | -0.648 |
| AsNAC042 | 145 | 442 | 9.46 | 16 896.43 | 38.54 | 62.48 | -0.587 |
| AsNAC043 | 149 | 454 | 9.46 | 17 444.08 | 36.77 | 63.42 | -0.533 |
| AsNAC044 | 130 | 396 | 9.80 | 15 114.11 | 16.76 | 50.23 | -0.893 |
| AsNAC045 | 149 | 454 | 9.39 | 17 409.75 | 35.18 | 66.04 | -0.799 |
| AsNAC046 | 140 | 426 | 9.43 | 16 452.80 | 46.91 | 63.36 | -0.699 |
| AsNAC047 | 251 | 765 | 5.24 | 28 836.27 | 31.96 | 54.74 | -0.734 |
| AsNAC048 | 260 | 792 | 5.04 | 29 891.58 | 47.35 | 94.42 | -0.202 |
| AsNAC049 | 261 | 796 | 8.63 | 29 843.57 | 37.33 | 67.59 | -0.593 |
表2 大蒜NAC蛋白的理化性质 (续表Continued)
Table 2 Physical and chemical properties of NAC proteins in Allium sativum L.
基因名称 Gene name | 氨基酸长度 Number of amino acid/aa | 编码区长度 Length of cDNA/bp | 等电点 Theoretical pI | 分子量 Molecular weight/Da | 不稳定指数 Instability index | 脂溶系数 Aliphatic index | 总平均疏水指数Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| AsNAC037 | 290 | 884 | 6.45 | 33 210.39 | 45.73 | 63.59 | -0.694 |
| AsNAC038 | 176 | 548 | 9.74 | 20 481.63 | 39.16 | 79.77 | -0.354 |
| AsNAC039 | 147 | 448 | 9.72 | 16 705.32 | 31.72 | 70.41 | -0.582 |
| AsNAC040 | 179 | 555 | 9.74 | 21 143.43 | 51.78 | 68.60 | -0.450 |
| AsNAC041 | 152 | 463 | 7.78 | 17 151.33 | 42.10 | 56.51 | -0.648 |
| AsNAC042 | 145 | 442 | 9.46 | 16 896.43 | 38.54 | 62.48 | -0.587 |
| AsNAC043 | 149 | 454 | 9.46 | 17 444.08 | 36.77 | 63.42 | -0.533 |
| AsNAC044 | 130 | 396 | 9.80 | 15 114.11 | 16.76 | 50.23 | -0.893 |
| AsNAC045 | 149 | 454 | 9.39 | 17 409.75 | 35.18 | 66.04 | -0.799 |
| AsNAC046 | 140 | 426 | 9.43 | 16 452.80 | 46.91 | 63.36 | -0.699 |
| AsNAC047 | 251 | 765 | 5.24 | 28 836.27 | 31.96 | 54.74 | -0.734 |
| AsNAC048 | 260 | 792 | 5.04 | 29 891.58 | 47.35 | 94.42 | -0.202 |
| AsNAC049 | 261 | 796 | 8.63 | 29 843.57 | 37.33 | 67.59 | -0.593 |
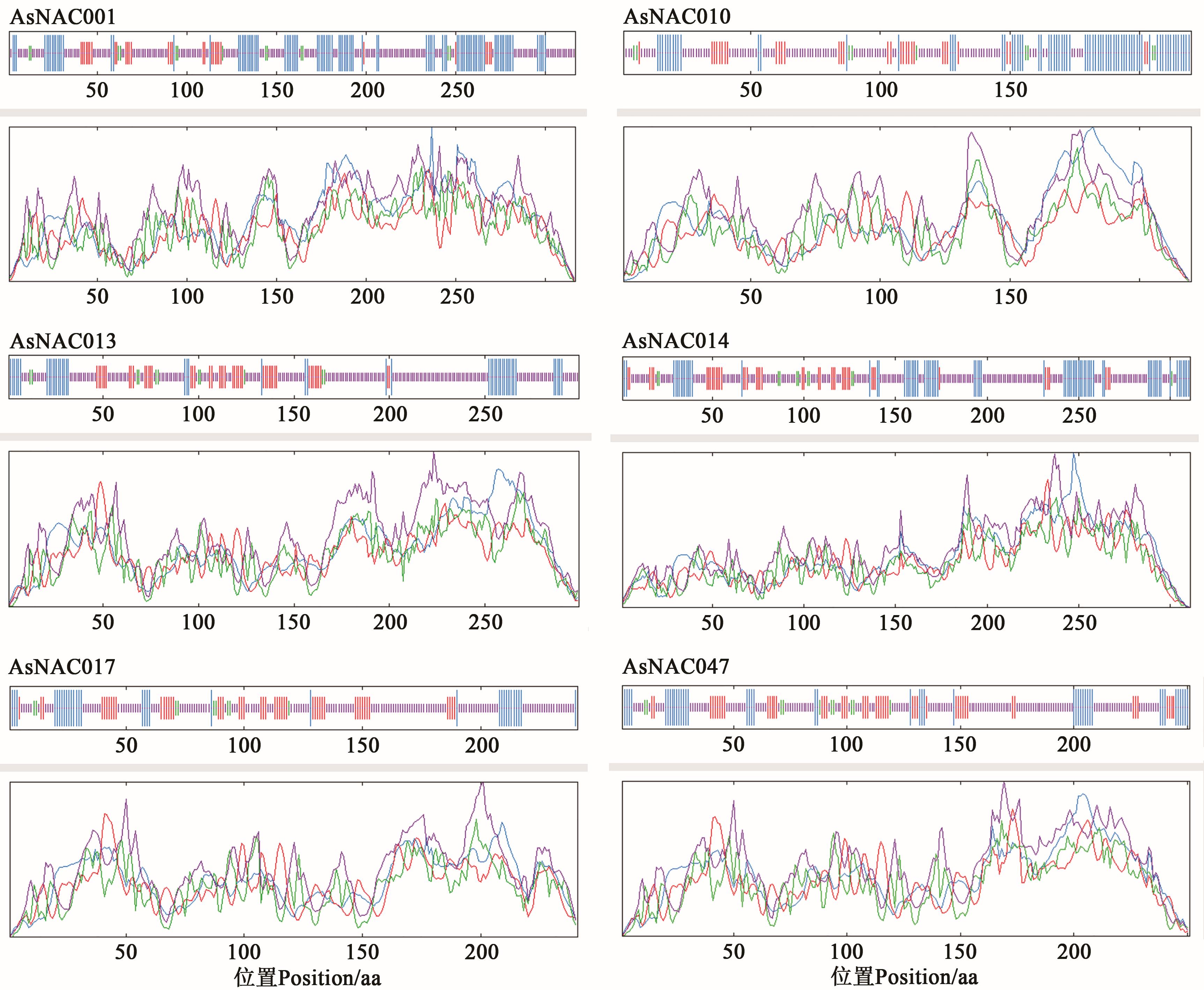
图3 大蒜NAC蛋白二级结构分析注:蓝色为α-螺旋;绿色为β-转角;红色为延长链;紫色为无规则卷曲。
Fig. 3 Secondary structure prediction of NAC proteins in Allium sativum L.Note: Blue indicates α-helix; green indicates β-turn; red indicates extended strand; purple indicates random curl.
编号 Code | α-螺旋 Alpha helix | 延长链 Extended strand | 无规卷曲 Random coil | β-转角 Beta turn |
|---|---|---|---|---|
| AsNAC001 | 31.55 | 9.46 | 54.89 | 4.10 |
| AsNAC010 | 32.73 | 15.00 | 48.18 | 4.09 |
| AsNAC013 | 16.05 | 15.38 | 64.88 | 3.68 |
| AsNAC014 | 24.44 | 13.83 | 58.20 | 3.54 |
| AsNAC017 | 13.69 | 20.33 | 62.24 | 3.73 |
| AsNAC047 | 17.53 | 19.52 | 58.96 | 3.98 |
表3 大蒜NAC蛋白二级结构分析 (%)
Table 3 Secondary structure prediction of NAC proteins in Allium sativum L.
编号 Code | α-螺旋 Alpha helix | 延长链 Extended strand | 无规卷曲 Random coil | β-转角 Beta turn |
|---|---|---|---|---|
| AsNAC001 | 31.55 | 9.46 | 54.89 | 4.10 |
| AsNAC010 | 32.73 | 15.00 | 48.18 | 4.09 |
| AsNAC013 | 16.05 | 15.38 | 64.88 | 3.68 |
| AsNAC014 | 24.44 | 13.83 | 58.20 | 3.54 |
| AsNAC017 | 13.69 | 20.33 | 62.24 | 3.73 |
| AsNAC047 | 17.53 | 19.52 | 58.96 | 3.98 |
| 1 | 王冰.国内外大蒜机械化播种与收获的分析与探讨[J].农业开发与装备,2020(3):159-161. |
| 2 | 赵洪光,滕文建,姚传峰.国内大蒜收获机械研制现状与对策[J].农业开发与装备,2018(12):80,82. |
| 3 | 李积善.青海省海东市乐都区现代富硒农业开发现状与发展对策[J].安徽农业科学,2018,46(33):217-220. |
| LI J S. Development status and countermeasures of modern selenium-rich agriculture in Ledu district of Haidong city in Qinghai province [J]. J. Anhui Agric. Sci., 2018, 46(33):217-220. | |
| 4 | 祝存锦.乐都区大蒜栽培技术和紫皮大蒜营养初探[J].现代农业,2017(5):46-47. |
| 5 | 吴新昌,陈剑雄,闫霞,等.大蒜生育期气候条件分析及种植区划[J].现代农业科技,2019(10):83-84, 86. |
| 6 | SOUE E, VAN HOUWELINGEN V, KLOOS D, et al.. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries [J]. Cell, 1996, 85(2):159-70. |
| 7 | 张磊,熊欢欢,曹庆,等.瞬时遗传转化长白落叶松NAC基因植株抗旱性的研究[J].植物研究,2020,40(3):394-400. |
| ZHANG L, XIONG H H, CAO Q, et al.. Drought resistance of larch NAC gene by transient genetic transformation [J]. Bot. Res., 2020, 40(3):394-400. | |
| 8 | 卢惠君,李子义,梁瀚予,等.刚毛柽柳NAC24基因的表达及抗逆功能分析[J].林业科学,2019,55(3):54-63. |
| LU H J, LI Z Y, LIANG H Y, et al.. Expression and stress tolerance analysis of NAC24 from Tamarix hispida [J]. Sci. Silv. Sin., 2019, 55(3):54-63. | |
| 9 | 段俊枝,李莹,赵明忠,等.NAC转录因子在水稻抗逆基因工程中的应用进展[J].中国稻米,2017,23(6):37-42, 46. |
| DUAN J Z, LI Y, ZHAO M Z, et al.. Progress on application of NAC transcription factors in rice stress tolerance genetic engineering [J]. China Rice, 2017, 23(6):37-42, 46. | |
| 10 | 张雪梅,姚文静,赵凯,等.杨树NAC7转录因子基因应答盐胁迫表达[J].东北林业大学学报,2017,45(8):6-9, 13. |
| ZHANG X M, YAO W J, ZHAO K, et al.. Expression analysis of NAC7 transcription factor gene from Populus simonii×P. nigra in response to salt stress [J]. J. Northeast For. Univ., 2017, 45(8): 6-9, 13. | |
| 11 | 张丽,张庭,谭登峰,等.玉米ZmNAC99基因的克隆及干旱诱导表达分析[J].西北植物学报,2017,37(4):629-635. |
| ZHANG L, ZHANG T, TAN D F, et al.. Isolation and drought induced expression characterization of ZmNAC99 gene from maize [J]. Acta Bot. Bor-Occid. Sin., 2017, 37(4):629-635. | |
| 12 | SUN Q G, JIANG S H, ZHANG T L, et al.. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB911 [J/OL]. Plant Sci., 2019, 289:110286 [2021-07-15]. . |
| 13 | HOU X M, ZHANG H F, LIU S Y, et al.. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers [J/OL]. Plant Sci., 2020, 291:110346 [2021-07-15]. . |
| 14 | ZHENG X, TANG S W, ZHU S Y, et al.. Identification of an NAC transcription factor family by deep transcriptome sequencing in onion (Allium cepa l.) [J/OL]. PloS One, 2016, 11(6): e0157871 [2021-07-15]. . |
| 15 | 赵艳青,杜建厂,王盼乔,等.哈氏黄瓜NAC转录因子的鉴定及低温表达分析[J].园艺学报,2019,46(7):1303-1319. |
| ZHAO Y Q, DU J C, WANG P Q, et al.. Identification and expression analysis of NAC transcription factor gene family under low temperature in Cucumis sativus var. [J]. Acta Hortic. Sin., 2019, 46(7):1303-1319. | |
| 16 | 宋琴,赵福宽,孙清鹏,等.洋葱AcNAC转录因子基因功能区的克隆与表达分析[C]//First International Conference on Cellular, molecular Biology, Biophysics & Bioengineering. 2010:417-422. |
| SONG Q, ZHAO F K, SUN Q P, et al.. Cloning and expression analysis of the functional region of the onion AcNAC transcription factor gene [C]//First International Conference on Cellular, molecular Biology, Biophysics & Bioengineering. 2010:417-422. | |
| 17 | 雍玉冰.低温胁迫应答转录因子在百合逆境响应中的功能及调控机制研究[D].北京:北京林业大学,2020. |
| YONG Y B. Function and transcriptional regulatory mechanism of cold stress responsive transcription factors in lily stress response [D]. Beijing: Beijing Forestry University, 2020. | |
| 18 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4):402-8. |
| 19 | 权宽章.乐都紫皮大蒜覆膜栽培技术[J].黑龙江农业科学,2009(3):169-170. |
| 20 | 周秉荣,胡爱军,陈国茜,等.青海省农牧业气候资源综合区划及评价[J].资源科学,2013(1):191-198. |
| 21 | YUAN P G, YANG T B, POOVAIAH B W. Calcium Signaling-mediated plant response to cold stress [J/OL]. Int. J. Mol. Sci., 2018, 19(12):3896 [2021-07-15]. . |
| 22 | KAZEMI-SHAHANDASHTI S S, MAALI-AMIRI R. Global insights of protein responses to cold stress in plants: signaling, defence and degradation [J]. J. Plant Physiol., 2018, 226: 123-135. |
| 23 | DOWD W W, KING F A, Denny M W. Thermal variation, thermal extremes and the physiological performance of individuals [J]. J. Exp. Biol.., 2015, 218(12):1956-1967. |
| 24 | ZHU T T, EVIATAR N, SUN D F, et al.. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants [J]. Evolution, 2012, 66(6):1833-1848. |
| 25 | HUANG D B, WANG S G, ZHANG B C, et al.. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice [J]. Plant Cell, 2015, 27(6):1681-1696. |
| 26 | MA X M, ZHANG Y J, TUREČKOVÁ V, et al.. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato [J]. Plant Physiol., 2018, 177(3):1286-1302. |
| 27 | PEI H X, MA N, TIAN J, et al.. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals [J]. Plant Physiol., 2013, 163(2):775-791. |
| 28 | 周鸿慧,黄红,徐彬磊,等.NAC转录因子在植物对生物和非生物胁迫响应中的功能[J].植物生理学报,2017,53(8):1372-1382. |
| ZHOU H H, HUANG H, XU B L, et al.. Biological function of NAC transcription factors in plant abiotic and biotic stress responses [J]. Acta Phytophysiol. Sin., 2017,53(8):1372-1382. | |
| 29 | LI X L, YANG X, HU Y X, et al.. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance [J]. Plant Cell Rep., 2014, 33(5):767-778. |
| 30 | MAO X G, CHEN S S, LI A, et al.. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis [J/OL]. PLoS One, 2017, 9(1): e84359 [2021-07-15]. . |
| 31 | 王营,关春景,崔颖,等.细叶百合LpNAC13基因的克隆及其表达[J].东北林业大学学报,2020,48(4):29-35. |
| WANG Y, GUAN C J, CUI Y, et al.. Cloning and expression analysis of LpNAC13 gene from Lilium pumilum . [J]. J. Northeast For. Univ., 2020, 48(4):29-35. | |
| 32 | OOKA H, SATOH K, DOI K, et al.. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana [J]. DNA Res., 2003, 10(6):239-247. |
| 33 | 王洋,柏锡.大豆NAC基因家族生物信息学分析[J].大豆科学,2014,33(3):325-333. |
| WANG Y, BAI X. Bioinformatics analysis of NAC gene family in Glycine max L. [J]. Soybean Sci., 2014, 33(3):325-333. | |
| 34 | 肖姗姗,赵虎,孙叶芳,等.小兰屿蝴蝶兰(Phalaenopsis equestris)NAC转录因子家族的全基因组序列鉴定及其进化分析[J].浙江农业学报,2016,28(7):1156-1163. |
| XIAO S S, ZHAO H, SUN Y F, et al.. Identification,characterization and evolution of NAC transcription factors in Phalaenopsis equestris [J]. Acta Agric. Zhejiangensis, 2016, 28(7):1156-1163. | |
| 35 | 姚丹,倪晓鹏,侍婷,等.果梅NAC基因家族的鉴定及组织表达分析[J].核农学报,2019,33(2):226-239. |
| YAO D, NI X P, SHI T, et al.. Identification and tissue expression analysis of NAC gene family in Prunus mume [J]. Acta Agric. Nucl. Sin., 2019, 33(2):226-239. | |
| 36 | FANG Y J, YOU J, XIE K B, et al.. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice [J]. Mol. Genet. Genomics, 2008, 280(6):547-563. |
| 37 | SUN H, HU M L, LI J Y, et al.. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton [J/OL]. BMC. Plant Biol., 2018, 18(1):150 [2021-07-15]. . |
| 38 | GONG X, ZHAO L Y, SONG X F, et al.. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri) [J/OL]. BMC. Plant Biol., 2019, 19(1):161 [2021-07-15]. . |
| 39 | ZHUO X K, ZHENG T C, ZHANG Z Y, et al.. Genome-wide analysis of the NAC transcription factor gene family reveals differential expression patterns and cold-stress responses in the woody plant Prunus mume [J]. Genes, 2018, 9(10):494-508. |
| [1] | 代祥, 宋海潮, 苏川. 自走式大蒜正芽播种施肥机设计与试验[J]. 中国农业科技导报, 2025, 27(9): 131-144. |
| [2] | 陈小伟, 沙玉柱, 刘秀, 邵鹏阳, 王翻兄, 谢转回, 杨文鑫, 陈倩玲, 高敏, 黄薇. 不同物候期藏绵羊肉品质、营养成分及肉质相关基因表达特征分析[J]. 中国农业科技导报, 2025, 27(7): 161-171. |
| [3] | 毛桃桃, 赵小强, 柏小栋, 余斌. 低温胁迫对玉米幼苗光合性能、抗氧化酶系统及相关基因表达的影响[J]. 中国农业科技导报, 2025, 27(5): 49-60. |
| [4] | 郭肖蓉, 刘颖, 樊佳珍, 黄涛, 周荣. 猪CREBRF基因生物信息学和表达规律分析[J]. 中国农业科技导报, 2024, 26(9): 44-53. |
| [5] | 顿国强, 王雷, 纪欣鑫, 姜新波, 赵宇, 郭娜. 金乡紫皮蒜种离散元参数标定与试验验证[J]. 中国农业科技导报, 2024, 26(8): 131-139. |
| [6] | 张绥林, 李洋, 李琰, 张赟齐, 齐建勋, 侯智霞. 核桃晚霜危害特性及影响机制研究进展[J]. 中国农业科技导报, 2024, 26(4): 18-26. |
| [7] | 表达特征分析, 姚亮伟, 沙玉柱, 郭新羽, 蒲小宁, 徐英, 王继卿, 李少斌, 郝志云, 刘秀. 不同海拔藏绵羊肉品质、营养成分及肉质相关基因[J]. 中国农业科技导报, 2024, 26(3): 66-75. |
| [8] | 苗宇, 王婕, 赵尧尧, 张丽佳, 刘美君. 低温胁迫后紫花苜蓿叶片光合作用的恢复特性研究[J]. 中国农业科技导报, 2024, 26(2): 80-89. |
| [9] | 刘化冰, 党伟, 李奇, 张晓兵, 徐志强, 钟永健, 任志广, 张勇刚, 袁凯龙, 杨浩, 王辉, 孙聚涛. 烟草品种间氮素吸收和同化差异研究[J]. 中国农业科技导报, 2024, 26(11): 66-78. |
| [10] | 钟鹏, 苗丽丽, 王建丽, 刘杰, 王晓龙. 油莎豆种质资源低温胁迫生理响应与耐寒性评价[J]. 中国农业科技导报, 2023, 25(9): 83-96. |
| [11] | 滕泽, 张玉霞, 陈卫东, 丛百明, 田永雷, 张庆昕, 张永亮, 王东儒. 壳聚糖对苜蓿抗寒性及抗寒保护物质含量的影响[J]. 中国农业科技导报, 2023, 25(2): 192-198. |
| [12] | 郭胜微, 边思文, 丁建文, 张晓辰, 杨兴, 杜锦, 向春阳. 糯玉米萌发期耐低温品种资源的综合评价[J]. 中国农业科技导报, 2023, 25(2): 38-47. |
| [13] | 黄丽芳, 龙宇宙, 李金芹, 董云萍, 王晓阳, 陈鹏, 王宪文, 闫林. 低温胁迫对小粒种咖啡幼苗生理特性的影响[J]. 中国农业科技导报, 2023, 25(2): 60-67. |
| [14] | 燕辉. 基于叶绿素荧光分析的油菜响应低温胁迫机制研究[J]. 中国农业科技导报, 2023, 25(1): 58-64. |
| [15] | 李夏夏, 张思语, 程智慧. 中国优质大蒜品种区域试验评价[J]. 中国农业科技导报, 2022, 24(7): 58-68. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
