




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (11): 83-92.DOI: 10.13304/j.nykjdb.2024.0333
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Rongju WU1( ), Nenggang CHEN1(
), Nenggang CHEN1( ), Huan LI1, Xiaoqing YAN1, Zhanlie YANG2, Jun LIU1
), Huan LI1, Xiaoqing YAN1, Zhanlie YANG2, Jun LIU1
Received:2024-04-25
Accepted:2024-06-05
Online:2025-11-15
Published:2025-11-17
Contact:
Nenggang CHEN
吴荣菊1( ), 陈能刚1(
), 陈能刚1( ), 李欢1, 鄢小青1, 杨占烈2, 刘军1
), 李欢1, 鄢小青1, 杨占烈2, 刘军1
通讯作者:
陈能刚
作者简介:吴荣菊 E-mail:rongju_wu@163.com;
基金资助:CLC Number:
Rongju WU, Nenggang CHEN, Huan LI, Xiaoqing YAN, Zhanlie YANG, Jun LIU. Genome-wide Identification and Bioinformatics Analysis of CesA/Csl Gene Family in Rice[J]. Journal of Agricultural Science and Technology, 2025, 27(11): 83-92.
吴荣菊, 陈能刚, 李欢, 鄢小青, 杨占烈, 刘军. 水稻CesA/Csl基因家族的全基因组鉴定及生物信息学分析[J]. 中国农业科技导报, 2025, 27(11): 83-92.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2024.0333
基因名称 Gene name | 基因号 Gene ID | 编码序列Coding sequence/bp | 氨基酸数量 Number of amino acids/aa | 分子量Molecular weight/kD | 等电点 pI | 亲水性系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| OsCesA1 | Os05g0176100 | 3 231 | 1 076 | 121.27 | 6.68 | -0.191 | 细胞膜 Cell membrane |
| OsCesA2 | Os03g0808100 | 3 222 | 1 073 | 120.68 | 7.86 | -0.214 | 细胞膜 Cell membrane |
| OsCesA3 | Os07g0424400 | 3 282 | 1 093 | 123.50 | 6.74 | -0.248 | 细胞膜 Cell membrane |
| OsCesA4 | Os01g0750300 | 2 970 | 989 | 111.03 | 6.09 | -0.112 | 细胞膜 Cell membrane |
| OsCesA5 | Os03g0837100 | 3 279 | 1 092 | 123.40 | 6.66 | -0.242 | 细胞膜 Cell membrane |
| OsCesA6 | Os07g0252400 | 3 279 | 1 092 | 122.41 | 7.60 | -0.211 | 细胞膜 Cell membrane |
| OsCesA7 | Os10g0467800 | 3 192 | 1 063 | 119.79 | 8.25 | -0.288 | 细胞膜 Cell membrane |
| OsCesA8 | Os07g0208500 | 3 246 | 1 081 | 120.79 | 8.03 | -0.217 | 细胞膜 Cell membrane |
| OsCesA9 | Os09g0422500 | 3 168 | 1 055 | 118.70 | 6.42 | -0.213 | 细胞膜 Cell membrane |
| OsCesA10 | Os12g0477200 | 1 017 | 338 | 37.46 | 9.53 | -0.339 | 高尔基体 Golgi apparatus |
| OsCesA11 | Os06g0601600 | 1 575 | 524 | 57.80 | 9.62 | -0.278 | 细胞膜 Cell membrane |
| OsCslD1 | Os10g0578200 | 774 | 257 | 28.55 | 9.10 | 0.750 | 高尔基体 Golgi apparatus |
| OsCslD2 | Os06g0111800 | 3 513 | 1 170 | 130.14 | 7.49 | -0.233 | 高尔基体 Golgi apparatus |
| OsCslD3 | Os08g0345500 | 3 441 | 1 146 | 127.15 | 6.48 | -0.227 | 高尔基体 Golgi apparatus |
| OsCslD4 | Os12g0555600 | 3 648 | 1 215 | 132.16 | 8.13 | -0.115 | 高尔基体 Golgi apparatus |
| OsCslD5 | Os06g0336500 | 3 039 | 1 012 | 112.40 | 8.07 | -0.144 | 高尔基体 Golgi apparatus |
| OsCslE1 | Os09g0478100 | 2 214 | 737 | 83.50 | 8.38 | -0.005 | 高尔基体 Golgi apparatus |
| OsCslE2 | Os02g0725300 | 2 238 | 745 | 83.65 | 7.69 | 0.042 | 高尔基体 Golgi apparatus |
| OsCslE3 | Os09g0478300 | 1 884 | 627 | 71.29 | 8.46 | -0.189 | 高尔基体 Golgi apparatus |
| OsCslH1 | Os10g0341700 | 2 253 | 750 | 83.83 | 6.93 | 0.062 | 高尔基体 Golgi apparatus |
| OsCslH2 | Os04g0429500 | 1 500 | 499 | 55.21 | 6.60 | 0.183 | 高尔基体 Golgi apparatus |
| OsCslH3 | Os04g0429600 | 2 379 | 792 | 88.49 | 8.43 | -0.114 | 高尔基体 Golgi apparatus |
Table 1 Basic physicochemical properties of proteins coded by CesA/Csl genes in Oryze sative
基因名称 Gene name | 基因号 Gene ID | 编码序列Coding sequence/bp | 氨基酸数量 Number of amino acids/aa | 分子量Molecular weight/kD | 等电点 pI | 亲水性系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| OsCesA1 | Os05g0176100 | 3 231 | 1 076 | 121.27 | 6.68 | -0.191 | 细胞膜 Cell membrane |
| OsCesA2 | Os03g0808100 | 3 222 | 1 073 | 120.68 | 7.86 | -0.214 | 细胞膜 Cell membrane |
| OsCesA3 | Os07g0424400 | 3 282 | 1 093 | 123.50 | 6.74 | -0.248 | 细胞膜 Cell membrane |
| OsCesA4 | Os01g0750300 | 2 970 | 989 | 111.03 | 6.09 | -0.112 | 细胞膜 Cell membrane |
| OsCesA5 | Os03g0837100 | 3 279 | 1 092 | 123.40 | 6.66 | -0.242 | 细胞膜 Cell membrane |
| OsCesA6 | Os07g0252400 | 3 279 | 1 092 | 122.41 | 7.60 | -0.211 | 细胞膜 Cell membrane |
| OsCesA7 | Os10g0467800 | 3 192 | 1 063 | 119.79 | 8.25 | -0.288 | 细胞膜 Cell membrane |
| OsCesA8 | Os07g0208500 | 3 246 | 1 081 | 120.79 | 8.03 | -0.217 | 细胞膜 Cell membrane |
| OsCesA9 | Os09g0422500 | 3 168 | 1 055 | 118.70 | 6.42 | -0.213 | 细胞膜 Cell membrane |
| OsCesA10 | Os12g0477200 | 1 017 | 338 | 37.46 | 9.53 | -0.339 | 高尔基体 Golgi apparatus |
| OsCesA11 | Os06g0601600 | 1 575 | 524 | 57.80 | 9.62 | -0.278 | 细胞膜 Cell membrane |
| OsCslD1 | Os10g0578200 | 774 | 257 | 28.55 | 9.10 | 0.750 | 高尔基体 Golgi apparatus |
| OsCslD2 | Os06g0111800 | 3 513 | 1 170 | 130.14 | 7.49 | -0.233 | 高尔基体 Golgi apparatus |
| OsCslD3 | Os08g0345500 | 3 441 | 1 146 | 127.15 | 6.48 | -0.227 | 高尔基体 Golgi apparatus |
| OsCslD4 | Os12g0555600 | 3 648 | 1 215 | 132.16 | 8.13 | -0.115 | 高尔基体 Golgi apparatus |
| OsCslD5 | Os06g0336500 | 3 039 | 1 012 | 112.40 | 8.07 | -0.144 | 高尔基体 Golgi apparatus |
| OsCslE1 | Os09g0478100 | 2 214 | 737 | 83.50 | 8.38 | -0.005 | 高尔基体 Golgi apparatus |
| OsCslE2 | Os02g0725300 | 2 238 | 745 | 83.65 | 7.69 | 0.042 | 高尔基体 Golgi apparatus |
| OsCslE3 | Os09g0478300 | 1 884 | 627 | 71.29 | 8.46 | -0.189 | 高尔基体 Golgi apparatus |
| OsCslH1 | Os10g0341700 | 2 253 | 750 | 83.83 | 6.93 | 0.062 | 高尔基体 Golgi apparatus |
| OsCslH2 | Os04g0429500 | 1 500 | 499 | 55.21 | 6.60 | 0.183 | 高尔基体 Golgi apparatus |
| OsCslH3 | Os04g0429600 | 2 379 | 792 | 88.49 | 8.43 | -0.114 | 高尔基体 Golgi apparatus |
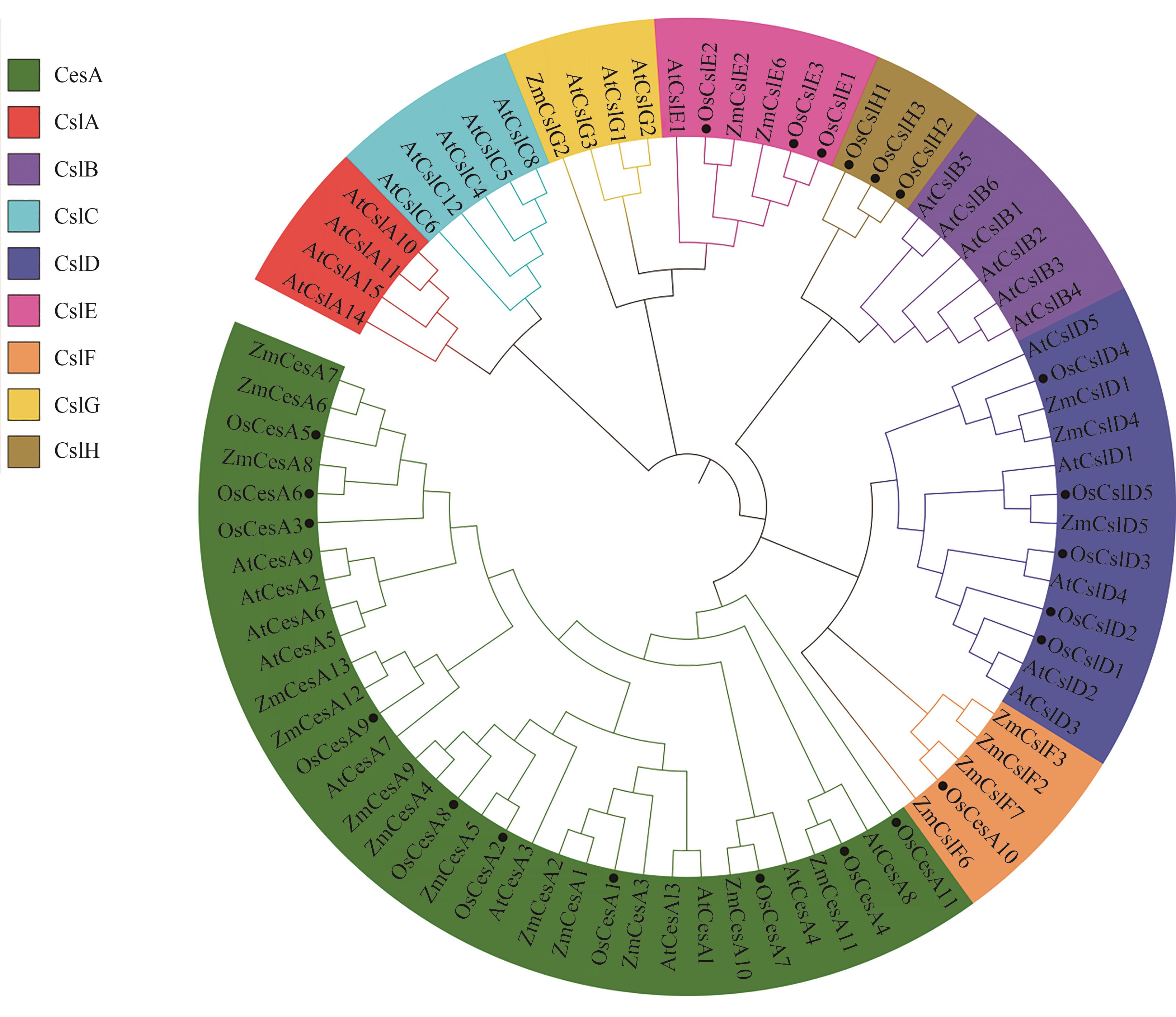
Fig. 2 Phylogenetic tree constructed by members of CesA/Csl protein in Oryze sative, Zea mays and Arabidopsis thalianaNote: Os—Oryze sative;Zm—Zea mays;At—Arabidopsis thaliana.
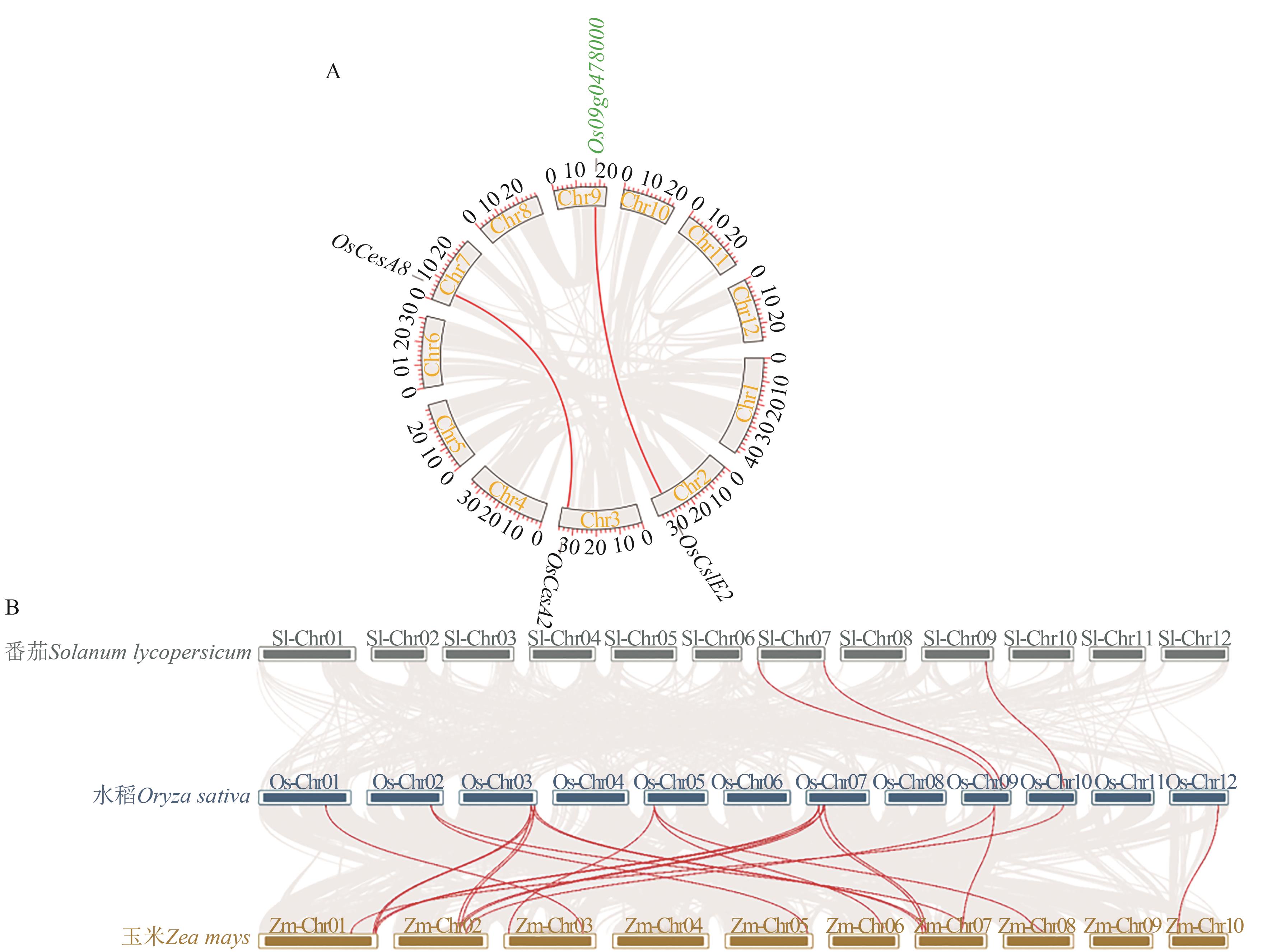
Fig. 5 Collinearity analysis of CesA/Csl genesA: Collinearity analysis within Oryze sative; B: Collinearity analysis among Oryze sative, Zea mays and Solanum lycopersicum
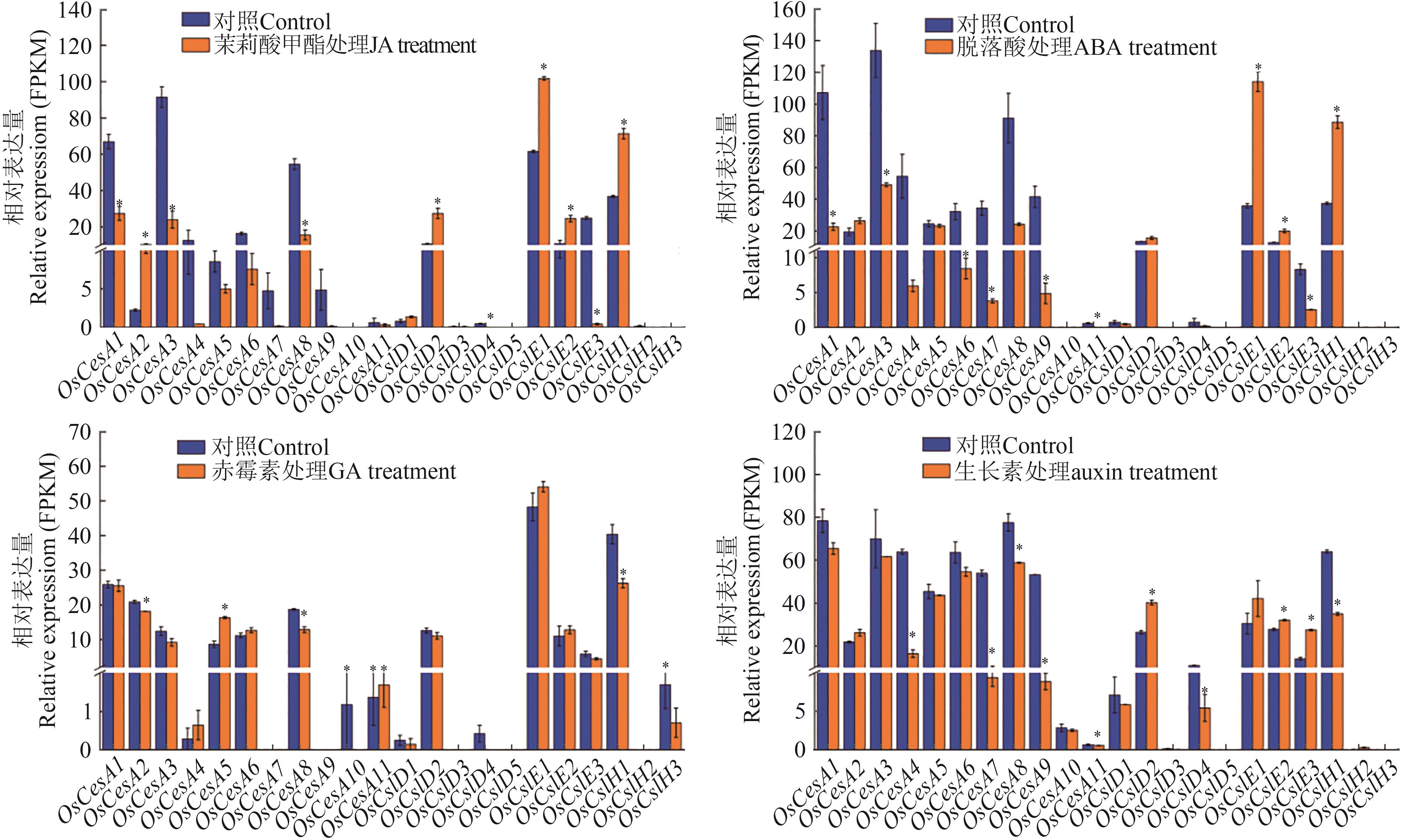
Fig. 7 Expression of CesA/Csl genes under different plant hormone treatmentsNote:* indicates significant difference between control and treatment at P<0.05 level.
| [1] | RICHMOND T A, SOMERVILLE C R. The cellulose synthase superfamily [J]. Plant Physiol., 2000, 124(2): 495-498. |
| [2] | TANAKA K, MURATA K, YAMAZAKI M, et al.. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall [J]. Plant Physiol., 2003, 133(1):73-83. |
| [3] | WANG D F, QIN Y L, FANG J J, et al.. A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice [J/OL]. PLoS One, 2016, 11(4):e0153993 [2024-03-20].. |
| [4] | WANG D F, YUAN S J, YIN L, et al.. A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice [J]. Plant Sci., 2012, 196:117-124. |
| [5] | LI F C, XIE G S, HUANG J F, et al.. OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice [J]. Plant Biotechnol. J., 2017, 15(9):1093-1104. |
| [6] | YU H, HU M, HU Z, et al.. Insights into pectin dominated enhancements for elimination of toxic Cd and dye coupled with ethanol production in desirable lignocelluloses [J/OL].Carbohydr. Polym., 2022, 286:119298 [2024-03-20]. . |
| [7] | HUANG D B, WANG S G, ZHANG B C, et al.. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice [J]. Plant Cell, 2015, 27(6):1681-1696. |
| [8] | KIM C M, PARK S H, JE B I, et al.. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice [J]. Plant Physiol., 2007, 143(3):1220-1230. |
| [9] | LI M, XIONG G Y, LI R, et al.. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth [J]. Plant J., 2009, 60(6):1055-1069. |
| [10] | LI R, XIONG G Y, ZHANG B C, et al.. Rice plants response to the disruption of OsCSLD4 gene [J]. Plant Signal. Behav., 2010,5(2):136-139. |
| [11] | HU J, ZHU L, ZENG D L, et al.. Identification and characterization of narrow androlled leaf 1, a novel gene regulating leaf morphology and plant architecture in rice [J]. Plant Mol. Biol., 2010, 73(3):283-292. |
| [12] | DING Z Q, LIN Z F, LI Q, et al.. DNL1, encodes cellulose synthase-like D4, is a major QTL for plant height and leaf width in rice (Oryza sativa L.) [J]. Biochem. Biophys. Res.Commun., 2015, 457(2):133-140. |
| [13] | QIAO L, WU Q L, YUAN L Z, et al.. SMALL PLANT AND ORGAN 1 (SPO1) encoding a cellulose synthase-like protein D4 (OsCSLD4) is an important regulator for plant architecture and organ size in rice [J/OL]. Int. J. Mol. Sci., 2023, 24(23):16974 [2024-03-20]. . |
| [14] | ZHAO H, LI Z X, WANG Y Y, et al.. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance [J]. Plant Biotechnol. J., 2022, 20(3):468-484. |
| [15] | LIU X, YIN Z L, WANG Y B, et al.. Rice cellulose synthase-like protein OsCSLD4 coordinates the trade-off between plant growth and defense [J/OL]. Front. Plant Sci., 2022, 13:980424 [2024-03-20]. . |
| [16] | CHEN C J, WU Y, LI J W, et al.. TBtools-Ⅱ:a “one for all, all for one” bioinformatics platform for biological big-data mining [J].Mol. Plant, 2023, 16(11): 1733-1742. |
| [17] | PANCALDI F, VAN LOO E N, SCHRANZ M E,et al.. Genomic architecture and evolution of the Cellulose synthase gene superfamily as revealed by phylogenomic analysis [J/OL].Front. Plant Sci., 2022, 13:870818 [2024-03-20].. |
| [18] | YIN Y, HUANG J, XU Y. The cellulose synthase superfamily in fully sequenced plants and algae [J/OL]. BMC Plant Biol., 2009, 9:99 [2024-03-20]. . |
| [19] | 田爱梅,许丽爱,陶贵荣,等.植物纤维素合酶[J].中国细胞生物学学报,2017,39(3):356-363. |
| TIAN A M, XU L A, TAO G R, et al.. Cellulose synthase in higher plants [J]. Chin. J. Cell Biol., 2017, 39(3):356-363. | |
| [20] | HAMANN T, OSBORNE E, YOUNGS H L, et al.. Global expression analysis of CESA and CSL genes in Arabidopsis [J].Cellulose, 2004, 11(3): 279-286. |
| [21] | DOBLIN M S, MELIS D L, NEWBIGIN E, et al.. Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families [J]. Plant Physiol., 2001, 125(4): 2040-2052. |
| [22] | KESTEN C, MENNA A, SÁNCHEZ-RODRÍGUEZ C. Regulation of cellulose synthesis in response to stress [J]. Curr. Opin. Plant Biol., 2017, 40: 106-113. |
| [23] | ELLIS C, TURNER J G. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens [J]. Plant Cell, 2001, 13(5): 1025-1033. |
| [24] | ELLIS C, KARAFYLLIDIS I, WASTERNACK C, et al.. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses [J]. Plant Cell, 2002, 14(7):1557-1566. |
| [1] | Jianfeng ZHANG, Wenfeng HOU, Yongqing WU, Kaixu LI, Xiaokun LI. Effects of Nitrogen Fertilizer and Density Interactions on Occurrence of Diseases and Insect Pests and Grain Yield of Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 145-154. |
| [2] | Zhien LIU, Yong HE, Zhicheng WANG, Xiaokang ZHAN, Tingbao WANG, Yaowei LIU, Zhihong TIAN. Identification and Bioinformatics Analysis of Growth Regulating Factor GRF Gene Family in Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 18-27. |
| [3] | Zhining YANG, Xuke LU, Yapeng FAN, Yuping SUN, Xin YU, Liang WANG, Yongjian GAO, Gulijiayinashen Habuli, Gulishaxi Nasiyi, Wumuer Abudu, Hui HUANG, Menghao ZHANG, Lidong WANG, Xiao CHEN, Lei XIAO, Xinrui ZHANG, Shuai WANG, Xiugui CHEN, Junjuan WANG, Lixue GUO, Wenwei GAO, Wuwei YE. Response Mechanism of Cotton GhDMT7 Gene to 5-azacytidine [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 28-35. |
| [4] | Zheng WU, Hongyun YANG, Aizhen SUN, Jie KONG, Shumei HUANG. Diagnosis of Potassium Nutrition in Rice Based on CA_MobileViT Model [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 80-88. |
| [5] | Lihua SHAO, Peng LI. Proteome Analysis of Rice Response to Gibberella fujikuroi Infection [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 126-135. |
| [6] | Wenting ZHANG, Yang LI, Shi QIU, Guangming LU, Dongshu GUO, Baolong ZHANG, Jinyan WANG. Effects of Badh2 Gene on Rice Quality Based on CRISPR/Cas9 Gene Editing Technology [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 39-48. |
| [7] | Yan WU, Leping ZOU, Huijie SONG, Dandan HU, Kailou LIU, Wanli LIANG. Effect of Controlled-release Nitrogen Fertilizer Combined Urea on Ammonium Nitrogen of Surface Water and Early Rice Yield [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 192-200. |
| [8] | Lintao CHEN, Zhaoxiang LIU, Ying LAN, Xiangwei MOU, Xu MA, Rijun WANG. Research on Rice Variety Identification Based on Hyperspectral Technology and Principal Component Analysis [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 104-111. |
| [9] | Lecheng SHEN, Zhigang WEN, Han LIAO, Xianbiao LIU, Yaocong JIANG, Yuancong ZHANG, Ting LIU, Mei WANG. Effects of Foliar Spraying of Different Selenium Fertilizers on Selenium Content, Selenium Speciation and Components in Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 206-215. |
| [10] | Dabing YANG, Liang HU, Xueshu DU, Bingliang WAN, Mingyuan XIA, Huaxiong QI, Jinbo LI. Progress in Creation of Rice Male Sterile Lines by CRISPR/Cas9 Gene Editing Technology [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 24-34. |
| [11] | Jinxian LIU, Lijuan WANG, Jie LIU, Xianyu FU, Guangheng WU. Identification and Expression Analysis of Calmodulin-binding Transcription Activator (CAMTA) Family Genes in Tea Plants [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 71-82. |
| [12] | Guoliang ZHONG, Lin WAN, Gang CHE, Hao TANG, Tianqi QU, Qilin ZHANG. Influence of Drying Parameters on Hot Air Drying Rate and Energy Consumption of Rice [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 95-103. |
| [13] | Lixia YI, Yong ZHOU, Wei YANG, Lai YAO, Mengdie JIANG, Jiangwen NIE, Bo ZHU, Zhangyong LIU. Effect of Ryegrass Return to Field as Substitute for Urea on Ammonia Volatilization from Paddy Soils [J]. Journal of Agricultural Science and Technology, 2025, 27(11): 186-194. |
| [14] | Pan ZHANG, Haichao SUN, Wenheng DONG, Yongjiang LI, Yingying ZHANG, Lili SHI, Jun ZHAO, Daowen LU. Genome-wide Identification and Expression Analysis of Maize JRL Gene Family [J]. Journal of Agricultural Science and Technology, 2025, 27(10): 32-41. |
| [15] | Qiujie CHEN, Yumei HUANG, Jinyan XIAN, Kaitong LI, Jielan HUANG, Xiaoxiu SHI, Liuping WANG, Dongping TU. Bioinformatics and Tissue-specific Expression Analysis of YUCCA Gene Family in Huperzia serrata [J]. Journal of Agricultural Science and Technology, 2025, 27(10): 72-83. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号