




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (6): 11-21.DOI: 10.13304/j.nykjdb.2024.0172
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Xinyue BAO1,2( ), Hongmin CHEN3(
), Hongmin CHEN3( ), Weiwei WANG2, Yimiao TANG2, Zhaofeng FANG2, Jinxiu MA2, Dezhou WANG2(
), Weiwei WANG2, Yimiao TANG2, Zhaofeng FANG2, Jinxiu MA2, Dezhou WANG2( ), Jinghong ZUO2(
), Jinghong ZUO2( ), Zhanjun YAO1(
), Zhanjun YAO1( )
)
Received:2024-03-07
Accepted:2024-03-26
Online:2024-06-15
Published:2024-06-12
Contact:
Dezhou WANG,Jinghong ZUO,Zhanjun YAO
鲍新跃1,2( ), 陈红敏3(
), 陈红敏3( ), 王伟伟2, 唐益苗2, 房兆峰2, 马锦绣2, 汪德州2(
), 王伟伟2, 唐益苗2, 房兆峰2, 马锦绣2, 汪德州2( ), 左静红2(
), 左静红2( ), 姚占军1(
), 姚占军1( )
)
通讯作者:
汪德州,左静红,姚占军
作者简介:鲍新跃E-mail:1772145342@qq.com基金资助:CLC Number:
Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes[J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21.
鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2024.0172
引物名称 Primer name | 正向引物序列 Forward primer sequence (3’-5’) | 反向引物序列 Reverse primer sequence (3’-5’) |
|---|---|---|
| TaCOBL-5A | GCGGCACCCGTGTCTTCTAT | CGTCTCGTCTCGTCGCAGTA |
| TaCOBL-5B | GCGGCACCCATGTCTTCTAT | CGTCTCGTCTCGTCGCAGTA |
| TaCOBL-5D | ACGGCACCCGCGTCTTCTAT | TCTCGTCGCTGTAAAAACTG |
| qTaCOBL-5A | CGTTGGATCTCTCTTGCAGC | TGGGATGGTCATGGGCAAAG |
| qTaCOBL-5B | GATTACGTGCAGGTTACATTCC | TCTCAAGGCTCCAGGTCAGG |
| qTaCOBL-5D | CAGCGAATCATAAGCCTCTG | GAGTAGCGGGGCAGGAAATG |
| TaActin | GGAATCCATGAGACCACCTAC | GACCCAGACAACTCGCAAC |
Table 1 Primers used in this study for gene cloning and qPCR
引物名称 Primer name | 正向引物序列 Forward primer sequence (3’-5’) | 反向引物序列 Reverse primer sequence (3’-5’) |
|---|---|---|
| TaCOBL-5A | GCGGCACCCGTGTCTTCTAT | CGTCTCGTCTCGTCGCAGTA |
| TaCOBL-5B | GCGGCACCCATGTCTTCTAT | CGTCTCGTCTCGTCGCAGTA |
| TaCOBL-5D | ACGGCACCCGCGTCTTCTAT | TCTCGTCGCTGTAAAAACTG |
| qTaCOBL-5A | CGTTGGATCTCTCTTGCAGC | TGGGATGGTCATGGGCAAAG |
| qTaCOBL-5B | GATTACGTGCAGGTTACATTCC | TCTCAAGGCTCCAGGTCAGG |
| qTaCOBL-5D | CAGCGAATCATAAGCCTCTG | GAGTAGCGGGGCAGGAAATG |
| TaActin | GGAATCCATGAGACCACCTAC | GACCCAGACAACTCGCAAC |
基因 Gene | 基因号 Gene ID number | 物理位置 Physical position/bp | 分子量 Molecular weight/Da | 等电点 pI | 蛋白长度 Protein length/aa | 预测定位 Predicted location |
|---|---|---|---|---|---|---|
| TaCOBL-5A | TraesCS5A02G392000 | 588 375 577~588 379 139 | 50 867.2 | 8.98 | 457 | 细胞膜Cell membrane |
| TaCOBL-5B | TraesCS5B02G396900 | 574 675 118~574 678 654 | 50 841.18 | 8.98 | 457 | 细胞膜Cell membrane |
| TaCOBL-5D | TraesCS5D02G401900 | 467 600 940~467 604 428 | 50 823.15 | 8.98 | 457 | 细胞膜Cell membrane |
Table. 2 TaCOBL-5 gene information and physicochemical properties analysis
基因 Gene | 基因号 Gene ID number | 物理位置 Physical position/bp | 分子量 Molecular weight/Da | 等电点 pI | 蛋白长度 Protein length/aa | 预测定位 Predicted location |
|---|---|---|---|---|---|---|
| TaCOBL-5A | TraesCS5A02G392000 | 588 375 577~588 379 139 | 50 867.2 | 8.98 | 457 | 细胞膜Cell membrane |
| TaCOBL-5B | TraesCS5B02G396900 | 574 675 118~574 678 654 | 50 841.18 | 8.98 | 457 | 细胞膜Cell membrane |
| TaCOBL-5D | TraesCS5D02G401900 | 467 600 940~467 604 428 | 50 823.15 | 8.98 | 457 | 细胞膜Cell membrane |
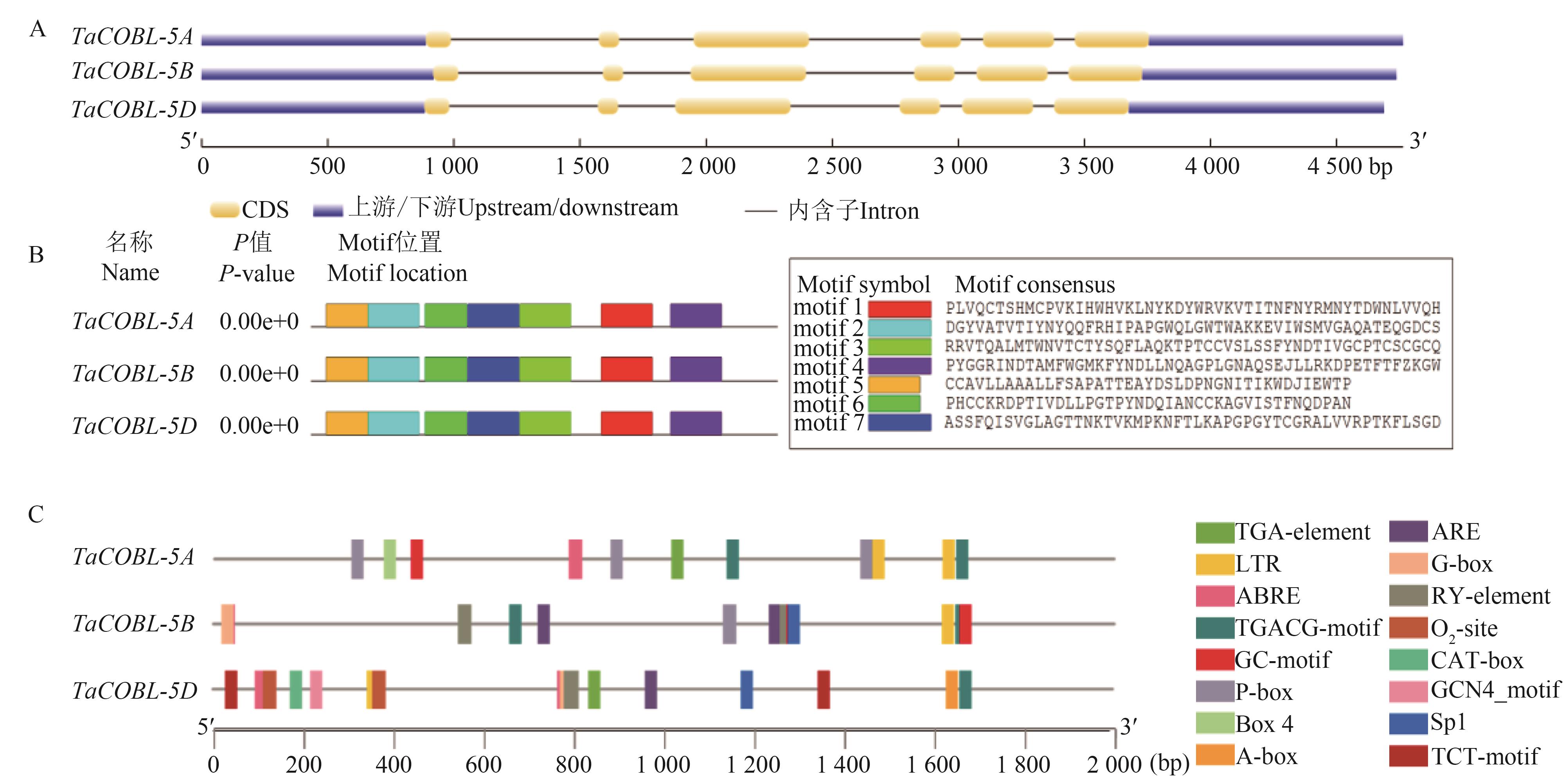
Fig. 1 Analysis of gene structure, conserved motif and cis-acting regulatory elements of TaCOBL-5A: Gene structure; B: Conserved motif; C: Cis-acting regulatory elements
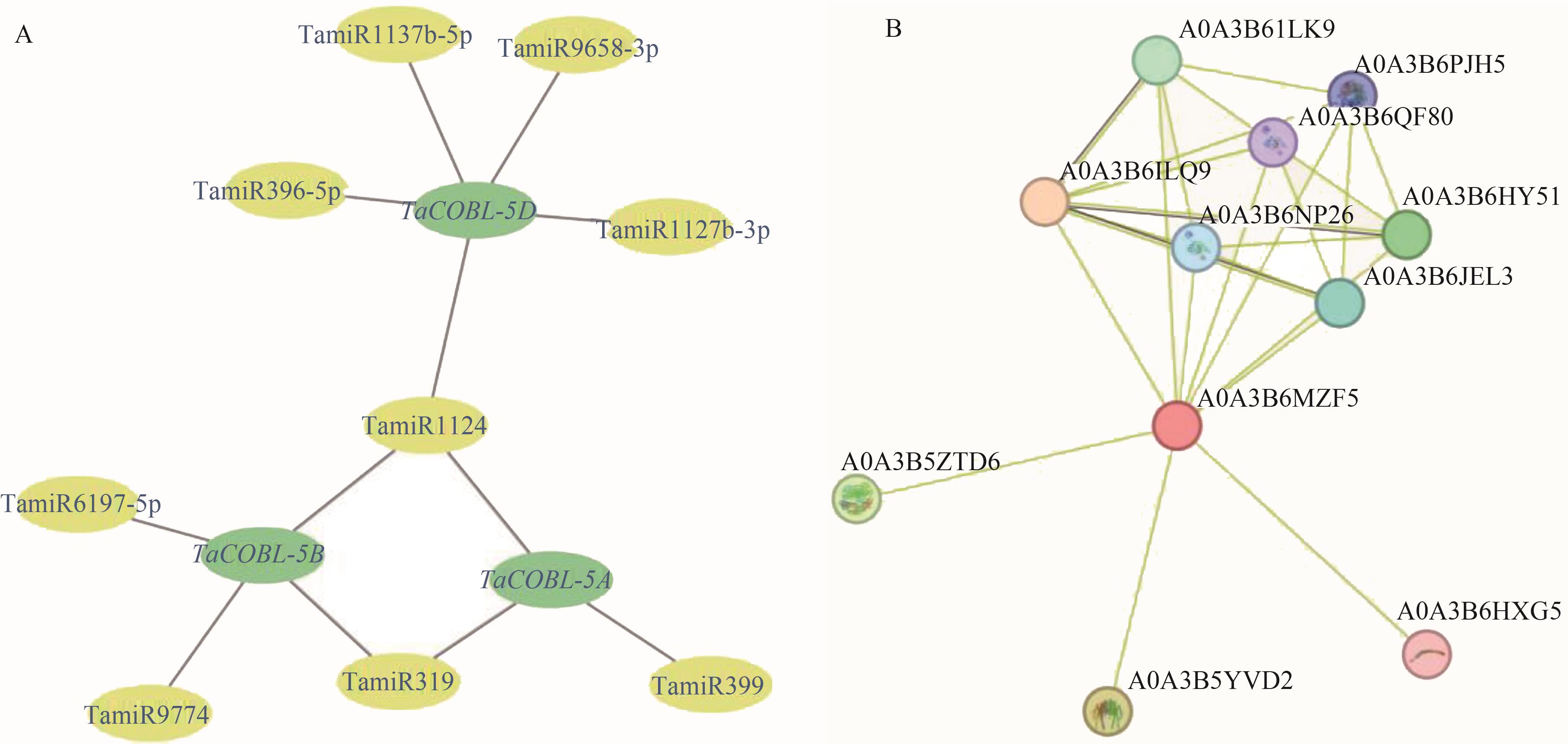
Fig. 5 Prediction putative networks of wheat miRNAs and the interacting proteinsA: Putative network of wheat miRNAs; B: Network of interacting proteins
| 1 | ZHU J K. Salt and drought stress signal transduction in plants [J]. Annu. Rev. Plant Biol., 2002, 53(1):247-273. |
| 2 | 陈翔,胡雨喆,陈甜甜,等.小麦抗低温逆境化控技术研究进展[J].植物营养与肥料学报, 2023, 29(8):1543-1555. |
| CHEN X, HU Y Z, CHEN T T, et al.. Progress of chemical regulation on wheat resistance to low temperature stress [J]. J. Plant Nutr. Fert., 2023, 29(8):1543-1555. | |
| 3 | WINFIELD M O, LU C G, WILSON I D, et al.. Plant responses to cold: transcriptome analysis of wheat [J]. Plant Biotechnol. J., 2010, 8(7):749-771. |
| 4 | MAO H D, JIAN C, CHENG X X, et al.. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency [J]. Plant Biotechnol. J., 2022, 20(5):846-861. |
| 5 | 温辉芹,程天灵,裴自友,等.山西中部区试小麦品种抗旱节水指标分析[J].山西农业科学,2020,48(10):1572-1575. |
| WEN H Q, CHENG T L, PEI Z Y, et al.. Analysis on drought resistance and water saving indexes of wheat varieties of regional trial in central Shanxi [J]. J. Shanxi Agric. Sci., 2020, 48(10):1572-1575. | |
| 6 | 健康,倪建平.植物非生物胁迫信号转导及应答[J].中国稻米, 2016, 22(6):52-60. |
| ZHU J K, NI J P. Abiotic stress signaling and responses in plants [J]. China Rice, 2016, 22(6):52-60. | |
| 7 | 盛松柏,田菊,庞晓明.冬枣COBRA基因家族全基因组鉴定及表达分析[J].分子植物育种, 2018, 16(1):61-68. |
| SHENG S B, TIAN J, PANG X M. Genome-wide identification and expression analysis of COBRA gene family in Ziziphus jujuba [J]. Mol. Plant Breeding, 2018, 16(1):61-68. | |
| 8 | SUN X M, XIONG H Y, JIANG C H, et al.. Natural variation of DROT1 confers drought adaptation in upland rice [J/OL]. Nat. Commun., 2022, 13(1):4265 [2024-04-03].. |
| 9 | LI Y H, QIAN O, ZHOU Y H, et al.. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants [J]. Plant Cell, 2003, 15(9):2020-2031. |
| 10 | BRADY S M, SONG S, DHUGGA K S, et al.. Combining expression and comparative evolutionary analysis. The COBRA gene family [J]. Plant Physiol., 2007, 143(1):172-187. |
| 11 | JULIUS B T, MCCUBBIN T J, MERTZ R A, et al.. Maize Brittle Stalk2-Like3, encoding a COBRA protein, functions in cell wall formation and carbohydrate partitioning [J]. Plant Cell, 2021, 33(10):3348-3366. |
| 12 | AHMED M Z, ALQAHTANI A S, NASR F A, et al.. Comprehensive analysis of the COBRA-like (COBL) gene family through whole-genome analysis of land plants [J]. Genet. Resour. Crop Evol., 2024, 71(1):863-872. |
| 13 | QIU C, CHEN J H, WU W H, et al.. Genome-wide analysis and abiotic stress-responsive patterns of COBRA-like gene family in Liriodendron chinense [J/OL]. Plants-Basel, 2023, 12(8):1616 [2024-04-03]. . |
| 14 | BEN-TOV D, ABRAHAM Y, STAV S, et al.. COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells [J]. Plant Physiol., 2015, 167(3):711-724. |
| 15 | LIU F F, WAN Y X, CAO W X, et al.. Novel function of a putative TaCOBL ortholog associated with cold response [J]. Mol. Biol. Rep., 2023, 50(5):4375-4384. |
| 16 | LIU L F, SHANG-GUAN K K, ZHANG B C, et al.. Brittle Culm1, a COBRA-Like protein, functions in cellulose assembly through binding cellulose microfibrils [J/OL]. PLoS Genet., 2013, 9(8):e1003704 [2024-04-03]. . |
| 17 | YE X, KANG B G, OSBURN D L, et al.. The COBRA gene family in Populus and gene expression in vegetative organs and in response to hormones and environmental stresses [J]. Plant Growth Regul., 2009, 58(2):211-223. |
| 18 | ZHANG D Q, YANG X H, ZHANG Z Y, et al.. Expression and nucleotide diversity of the poplar COBL gene [J]. Tree Genet. Genomes, 2010, 6(2):331-344. |
| 19 | YILAN E, XIN G, JING X, et al.. Genome-wide identification of the COBRA-like gene family in Pinus tabuliformis and the role of PtCOBL12 in the regulation of cellulose biosynthesis [J]. Ind. Crop Prod., 2023, 203. |
| 20 | YANG Q, WANG S, CHEN H, et al.. Genome-wide identification and expression profiling of the COBRA-like genes reveal likely roles in stem strength in rapeseed (Brassica napus L.) [J/OL]. PLoS One, 2021,16(11):e0260268 [2024-04-03]. . |
| 21 | NIU E L, SHANG X G, CHENG C Z, et al.. Comprehensive analysis of the COBRA-Like (COBL) gene family in Gossypium identifies two COBLs potentially associated with fiber quality [J/OL]. PLoS One, 2015, 10(12):e014572 [2024-04-03]. . |
| 22 | SANGI S, ARAÚJO PAULA M, COELHO F S, et al.. Genome-wide analysis of the COBRA-Like gene family supports gene expansion through whole-genome duplication in soybean (Glycine max) [J]. Plants, 2021, 10 (1):167-167. |
| 23 | XU L, WANG D Z, LIU S, et al.. Comprehensive atlas of wheat (Triticum aestivum L.) AUXIN RESPONSE FACTOR expression during male reproductive development and abiotic stress [J/OL]. Front. Plant Sci., 2020, 11:586144 [2024-04-03]. . |
| 24 | 吴凯铭.337份小麦品种(系)萌发期抗旱性鉴定及生理响应[D].杨凌:西北农林科技大学,2022. |
| WU K M. Identification of drought resistance and physiological response during germination in 337 wheat varieties (lines) [D]. Yangling: Northwest A&F University, 2022. | |
| 25 | 史冰新.黄淮和长江中下游冬麦区小麦耐热种质资源筛选[D].杨凌:西北农林科技大学,2023. |
| SHI B X. Screened for heat tolerance of wheat germplasm resources in the Yellow and Huai River valley winter wheat zone and the middle and lower Yangtze valley winter wheat zone [D]. Yangling: Northwest A&F University, 2023. | |
| 26 | KESAWAT M S, KHERAWAT B S, SINGH A, et al.. Genome-wide identification and characterization of the Brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.) [J]. Int. J. Mol. Sci., 2021, 22(16):8743-8743. |
| 27 | FANG Y J, ZHENG Y Q, LU W, et al.. Roles of miR319-regulated TCPs in plant development and response to abiotic stress [J]. Crop J., 2021, 9(1):17-28. |
| 28 | JIAN C, HAO P A, HAO C Y, et al.. The miR319/TaGAMYB3 module regulates plant architecture and improves grain yield in common wheat (Triticum aestivum) [J]. New Phytol., 2022, 235(4):1515-1530. |
| 29 | 梁婷,左静红,陆青,等.小麦IQM基因家族鉴定及非生物胁迫下表达分析[J].中国农业科技导报, 2023, 25(2):27-37. |
| LIANG T, LU Q, ZUO J Het al.. Identification and expression analysis under abiotic stress of IQM gene family in wheat (Triticum aestivum L.) [J]. J. Agric. Sci. Technol., 2023, 25(2):27-37. | |
| 30 | 陆青,梁婷,汪德州,等.小麦热激蛋白基因TaHSP90-1的克隆与表达分析[J].中国农业科技导报, 2022, 24(8):44-54. |
| LU Q, LIANG T, WANG D Zet al.. Cloning and expression analysis of wheat heat shock protein gene TaHSP90-1 [J]. J. Agric. Sci. Technol., 2022, 24(8):44-54. |
| [1] | Yifan LIU, Shaoshuai LIU, Rui ZANG, Yang LI, Wei LIU, Tingting LI, Danmei LIU, Dengcai LIU, Aili LI, Long MAO, Xiang WANG, Shuaifeng GENG. Analysis of Quality Traits of 168 Wheat Germplasm Resources [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 44-57. |
| [2] | Caixia LYU, Yongfu LI, Huinan XIN, Na LI, Ning LAI, Qinglong GENG, Shuhuang CHEN. Effects of Slow Release Nitrogen Fertilizer on Yield of Winter Wheat and Soil Nitrate/Ammonium Nitrogen Under Drip Irrigation [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 179-186. |
| [3] | Zhining YANG, Xuke LU, Yapeng FAN, Yuping SUN, Xin YU, Liang WANG, Yongjian GAO, Gulijiayinashen Habuli, Gulishaxi Nasiyi, Wumuer Abudu, Hui HUANG, Menghao ZHANG, Lidong WANG, Xiao CHEN, Lei XIAO, Xinrui ZHANG, Shuai WANG, Xiugui CHEN, Junjuan WANG, Lixue GUO, Wenwei GAO, Wuwei YE. Response Mechanism of Cotton GhDMT7 Gene to 5-azacytidine [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 28-35. |
| [4] | Qiang ZHU, Zongxian CHE, Heng CUI, Jiudong ZHANG, Xingguo BAO. Effect of Green Manure Replacing Nitrogen Fertilizer on Greenhouse Gases in Wheat Fields [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 182-189. |
| [5] | Yi HU, Jie GONG, Wei ZHAO, Rong CHENG, Zhongyu LIU, Shiqing GAO, Yazhen YANG. Identification of PHY Gene Family in Wheat (Triticum aestivum L.)and Their Expression Analysis Under Heat Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 30-43. |
| [6] | Jilin SUN, Jiaqi ZHANG, Fansen MENG, Silong SUN. Genome-wide Identification and Tissue Expression Pattern Analysis of HSP70 Gene Family in Thinopyrum elongatum [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 28-38. |
| [7] | Sile HU, Yulong BAO, Tubuxinbayaer, Jifeng TAO, Enliang GUO. Chlorophyll Content Inversion of Spring Wheat Based on Unmanned Aerial Vehicle Hyperspectral and Integrated Learning [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 93-103. |
| [8] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [9] | Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35. |
| [10] | Yixin CHEN, Xiubo YANG, Shijun TIAN, Cong WANG, Zhiying BAI, Cundong LI, Ke ZHANG. Response of GhCOMT28 to Drought Stress in Gossypium hirsutum [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 45-56. |
| [11] | Jinxian LIU, Lijuan WANG, Jie LIU, Xianyu FU, Guangheng WU. Identification and Expression Analysis of Calmodulin-binding Transcription Activator (CAMTA) Family Genes in Tea Plants [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 71-82. |
| [12] | Zhenyu XUE, Kangkang ZHANG, Yuanyuan ZHANG, Qiangqiang YAN, Lirong YAO, Hong ZHANG, Yaxiong MENG, Erjing SI, Baochun LI, Xiaole MA, Huajun WANG, Juncheng WANG. Screening and Functional Gene Detection of High-quality and Drought-resistant Wheat Germplasms [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 35-49. |
| [13] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [14] | Xianyin SUN, Qiuhuan MU, Yong MI, Guangde LYU, Xiaolei QI, Yingying SUN, Xundong YIN, Ruixia WANG, Ke WU, Zhaoguo QIAN, Yan ZHAO, Minggang GAO. Classification and Evaluation of New Wheat Lines Based on GT Biplot [J]. Journal of Agricultural Science and Technology, 2024, 26(7): 14-24. |
| [15] | Yangyang DU, Yuanyuan BAO, Xiangyu LIU, Xinyong ZHANG. Effects of Tartary Buckwheat Rotation on Enzyme Activities and Microorganisms in Rhizosphere Soil of Cultivated Potato in Yunnan Province [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 192-200. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号