




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (11): 226-239.DOI: 10.13304/j.nykjdb.2025.0240
• INNOVATIVE METHODS AND TECHNOLOGIES • Previous Articles
Hao XIE1,2( ), Zhilong CHEN2,3,4,5, Cuiting PENG2,3, Yingting PAN2,3, Lin QI2,3, Yulan ZHAO2,3, Yinglian BI6, Ziyi SONG1(
), Zhilong CHEN2,3,4,5, Cuiting PENG2,3, Yingting PAN2,3, Lin QI2,3, Yulan ZHAO2,3, Yinglian BI6, Ziyi SONG1( ), Zhonglin TANG2,3,4,5(
), Zhonglin TANG2,3,4,5( )
)
Received:2025-04-04
Accepted:2025-05-26
Online:2025-11-15
Published:2025-11-17
Contact:
Ziyi SONG,Zhonglin TANG
谢浩1,2( ), 陈指龙2,3,4,5, 彭翠婷2,3, 潘颖婷2,3, 齐霖2,3, 赵玉兰2,3, 闭英连6, 宋子仪1(
), 陈指龙2,3,4,5, 彭翠婷2,3, 潘颖婷2,3, 齐霖2,3, 赵玉兰2,3, 闭英连6, 宋子仪1( ), 唐中林2,3,4,5(
), 唐中林2,3,4,5( )
)
通讯作者:
宋子仪,唐中林
作者简介:谢浩 E-mail:xiehao2024888@163.com
基金资助:CLC Number:
Hao XIE, Zhilong CHEN, Cuiting PENG, Yingting PAN, Lin QI, Yulan ZHAO, Yinglian BI, Ziyi SONG, Zhonglin TANG. Preparation of Embryos with Simultaneous Editing of MSTN, pAPN and CD163 Genes in Pig[J]. Journal of Agricultural Science and Technology, 2025, 27(11): 226-239.
谢浩, 陈指龙, 彭翠婷, 潘颖婷, 齐霖, 赵玉兰, 闭英连, 宋子仪, 唐中林. 猪MSTN、pAPN和CD163基因同步编辑的胚胎制备[J]. 中国农业科技导报, 2025, 27(11): 226-239.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2025.0240
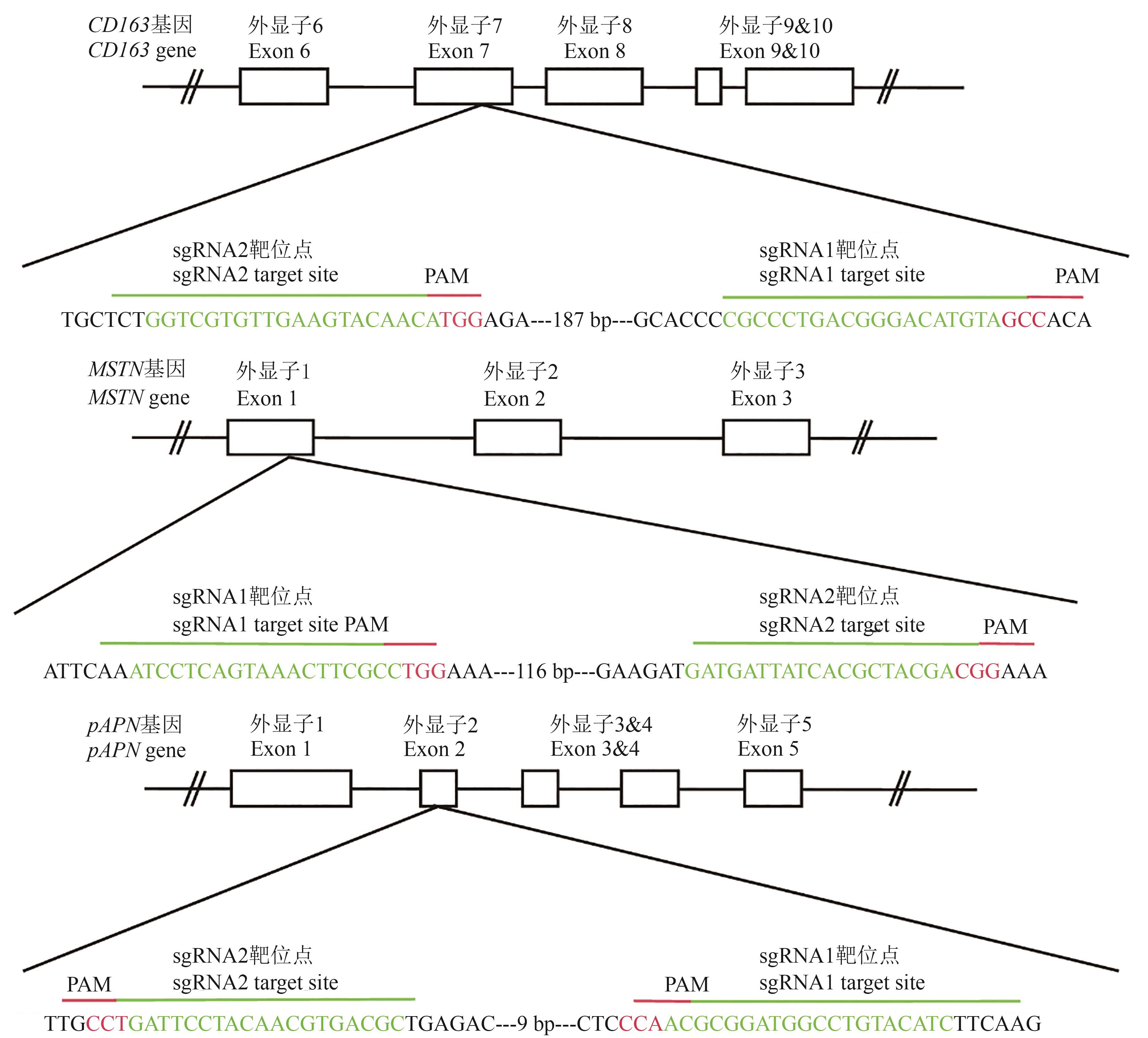
Fig. 1 sgRNA positions and sequences of MSTN, CD163 and pAPN genesNote:The box indicates exon; green indicates sgRNA sequence; red indicates PAM sequences.
| 引物名称Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| CD163-sgRNA1 | F: | R: |
| CD163-sgRNA2 | F: | R: |
| MSTN-sgRNA1 | F: | R: |
| MSTN-sgRNA2 | F: | R: |
| pAPN-sgRNA1 | F: | R: |
| pAPN-sgRNA2 | F: | R: |
Table 1 Oligo nucleotide sequence of sgRNA
| 引物名称Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| CD163-sgRNA1 | F: | R: |
| CD163-sgRNA2 | F: | R: |
| MSTN-sgRNA1 | F: | R: |
| MSTN-sgRNA2 | F: | R: |
| pAPN-sgRNA1 | F: | R: |
| pAPN-sgRNA2 | F: | R: |
引物名称 Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| 上游Forward | 下游Reverse | |
| CD163-ko | ATGGGTTCCAGAAGGCAAAG | CCATTCACCAAGCGGATTT |
| pAPN-ko | TGTCTGAGCCCTGGTTAATTT | TTGAGCTTCTTGCTATGGATG |
| MSTN-ko | TGAATGAGAACAGCGAGCAA | ATGCCTATTTCAGACAACCAAC |
| ucs-homo | acaaatggctctagaggtaccGAGGGCCTATTTCCCATGATTC | atcatgggaaataggccctcGCACCGACTCGGTGCCAC |
| uas-homo | GAGGGCCTATTTCCCATGATTC | gtaagttatgtaacgggtaccGCACCGACTCGGTGCCAC |
| MSTNsg1-OT-1 | TGCAAAGCTGGACCCACAAAG | AGCTGCAGATGCTCACCTG |
| MSTNsg1-OT-2 | GTAGTGTAGGCCAGCAGCTCTA | ATCTCCAGAGAATCAGGCTGA |
| MSTNsg1-OT-3 | CTTTTCCGCCAAAGCTGTTT | GTTCTCAGACCATGACTATGG |
| CD163sg1-OT-1 | GGTCTCAAACGTCTCCCCT | ACCCGCTCTCCCCCTTCTC |
| CD163sg1-OT-2 | GTTCCCAGGACTGGAGAGG | GTGTCCCTGCTCCCCAGG |
| CD163sg1-OT-3 | TTCCTGACCACCCCACCC | GCCACTGCAACACCAGATCT |
| pAPNsg1-OT-1 | GTGATTTCCCGAAGCCTGTT | GGCTGGGGGTTCCTTCCT |
| pAPNsg1-OT-2 | AGGCTTCCGGAAAATTAAGCTA | GGAGCAGATTAACAGAGACC |
| pAPNsg1-OT-3 | GGAGCAAGTGTGGTACCATG | TTGGAAGATTCACAACTGTAGA |
Table 2 Primer sequence
引物名称 Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| 上游Forward | 下游Reverse | |
| CD163-ko | ATGGGTTCCAGAAGGCAAAG | CCATTCACCAAGCGGATTT |
| pAPN-ko | TGTCTGAGCCCTGGTTAATTT | TTGAGCTTCTTGCTATGGATG |
| MSTN-ko | TGAATGAGAACAGCGAGCAA | ATGCCTATTTCAGACAACCAAC |
| ucs-homo | acaaatggctctagaggtaccGAGGGCCTATTTCCCATGATTC | atcatgggaaataggccctcGCACCGACTCGGTGCCAC |
| uas-homo | GAGGGCCTATTTCCCATGATTC | gtaagttatgtaacgggtaccGCACCGACTCGGTGCCAC |
| MSTNsg1-OT-1 | TGCAAAGCTGGACCCACAAAG | AGCTGCAGATGCTCACCTG |
| MSTNsg1-OT-2 | GTAGTGTAGGCCAGCAGCTCTA | ATCTCCAGAGAATCAGGCTGA |
| MSTNsg1-OT-3 | CTTTTCCGCCAAAGCTGTTT | GTTCTCAGACCATGACTATGG |
| CD163sg1-OT-1 | GGTCTCAAACGTCTCCCCT | ACCCGCTCTCCCCCTTCTC |
| CD163sg1-OT-2 | GTTCCCAGGACTGGAGAGG | GTGTCCCTGCTCCCCAGG |
| CD163sg1-OT-3 | TTCCTGACCACCCCACCC | GCCACTGCAACACCAGATCT |
| pAPNsg1-OT-1 | GTGATTTCCCGAAGCCTGTT | GGCTGGGGGTTCCTTCCT |
| pAPNsg1-OT-2 | AGGCTTCCGGAAAATTAAGCTA | GGAGCAGATTAACAGAGACC |
| pAPNsg1-OT-3 | GGAGCAAGTGTGGTACCATG | TTGGAAGATTCACAACTGTAGA |
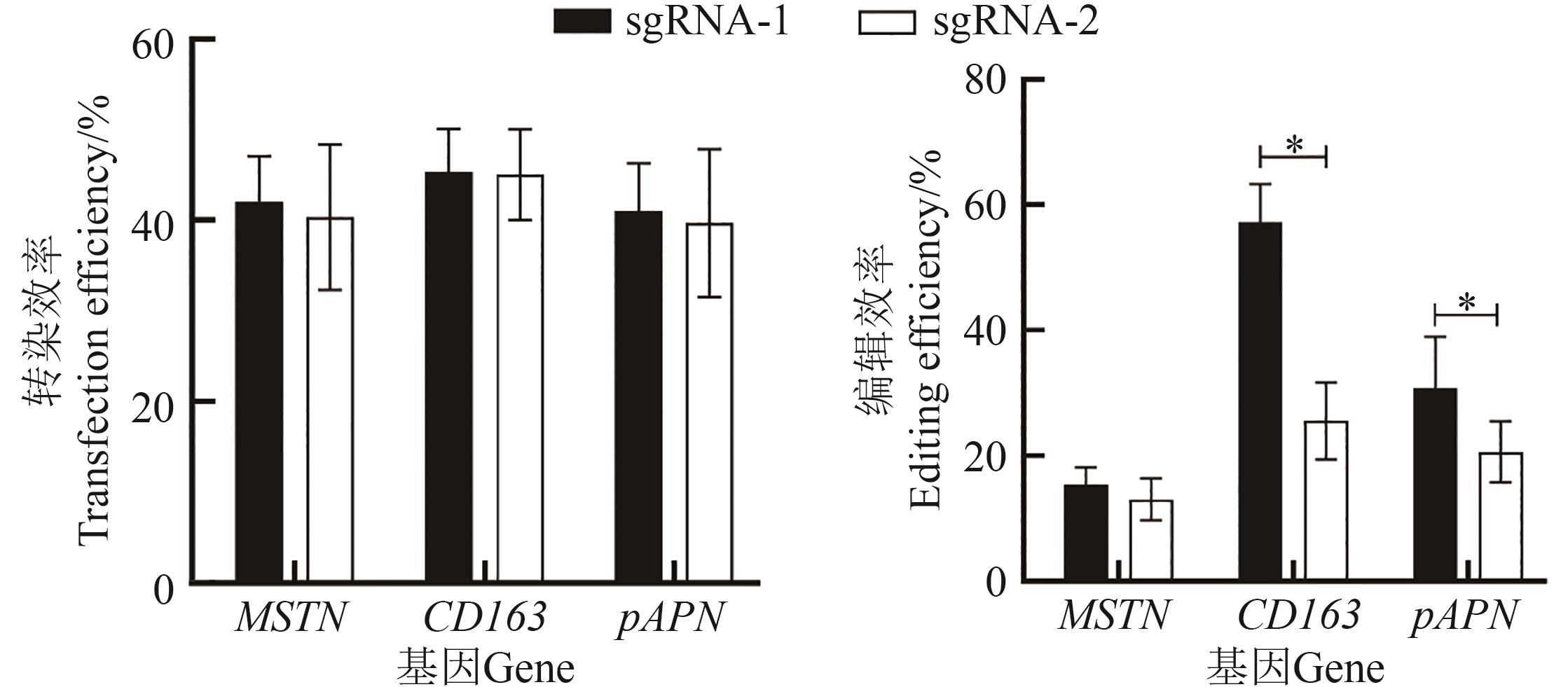
Fig. 3 Transfection and editing efficiency of different sgRNAs targeting MSTN, CD163 and pAPN genesNote:* indicates significant difference at P<0.05 level.
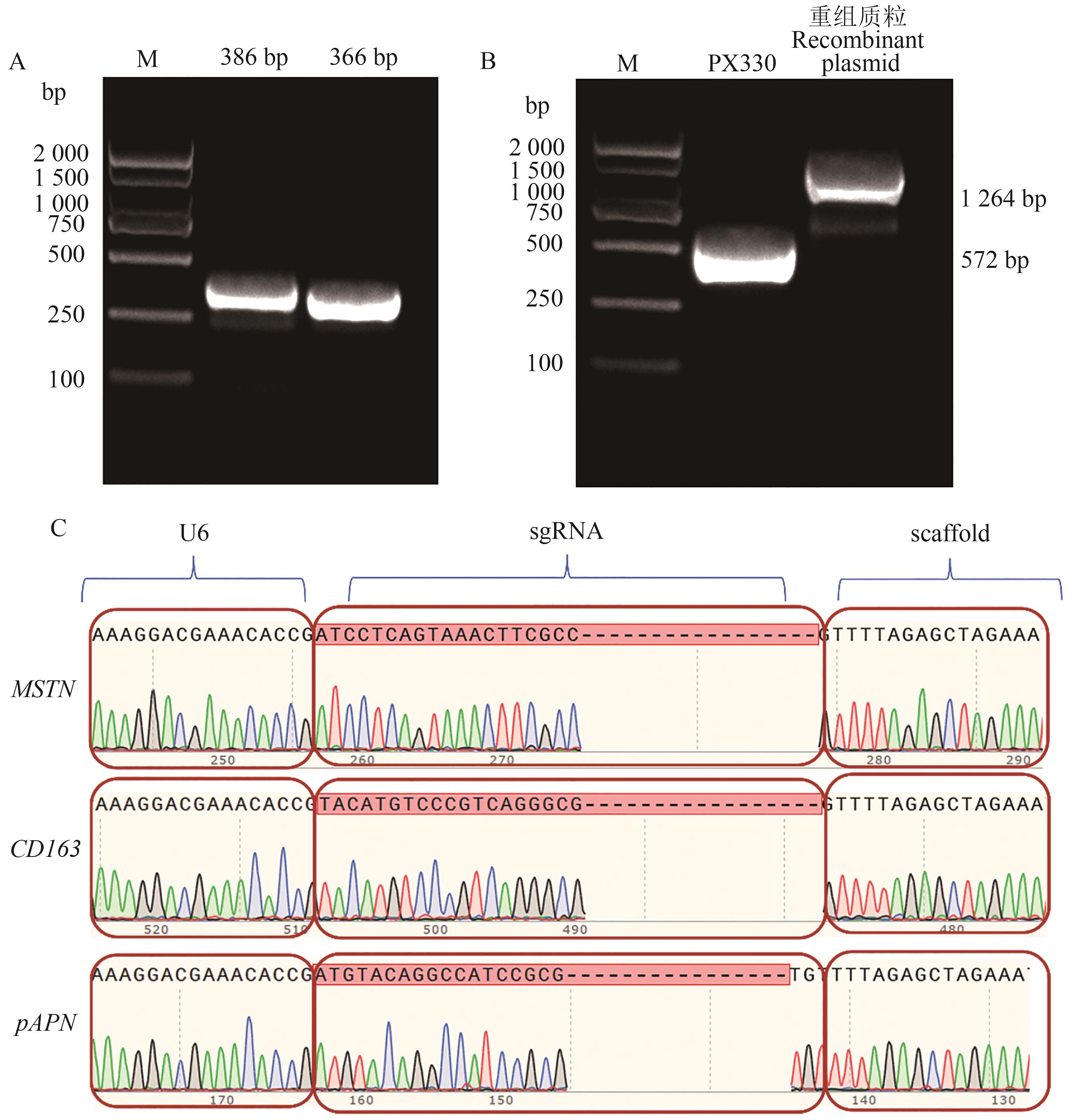
Fig. 4 Identification of multi-gene editing plasmid with MSTN, CD163 and pAPN genesA: Products of amplified fragments; B: PCR of recombinant plasmid; C: Ssequencing of multi-gene editing plasmid. M—2 000 bp DNA Marker

Fig. 6 Deletion sequences of 3 gene in knock out cellNote:1#~45# are different monoclones; the green font indicates the original sequence; the red font indicates the substitutions; the orange font indicates the insertions; the red dashes indicates the deletions.
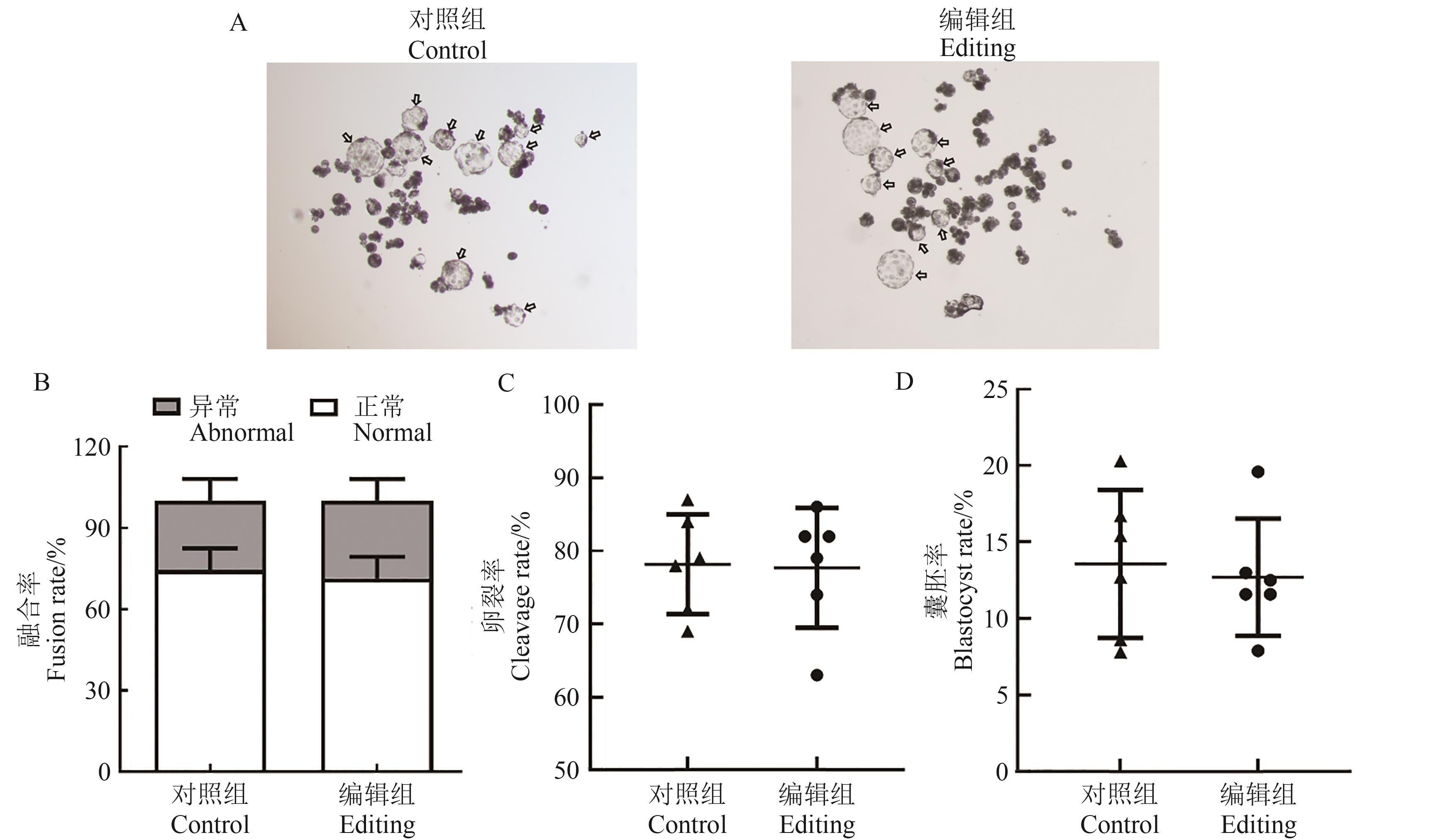
Fig. 7 Verification of multi-gene editing of MSTN, CD163 and pAPN at embryonic levelA: Representative images of blastocysts in each group, the arrows indicate blastocysts; B: Somatic cell fusion rate; C: Embryo cleavage rate; D: Blastocyst formation rate in each group
| [1] | LIU W, LI L, JIANG J, et al.. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics [J]. Precis. Clin. Med., 2021, 4(3): 179-191. |
| [2] | CHOJNACKA-PUCHTA L, SAWICKA D. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications [J]. J. Appl. Genet., 2020, 61(2): 221-229. |
| [3] | WHYTE J J, PRATHER R S. Genetic modifications of pigs for medicine and agriculture [J]. Mol. Reprod. Dev., 2011, 78(10-11): 879-891. |
| [4] | BIBIKOVA M, BEUMER K, TRAUTMAN J K, et al.. Enhancing gene targeting with designed zinc finger nucleases [J/OL]. Science, 2003, 300(5620): 764 [2025-03-20]. . |
| [5] | MILLER J C, TAN S, QIAO G, et al.. A TALE nuclease architecture for efficient genome editing [J]. Nat. Biotechnol., 2011, 29(2): 143-148. |
| [6] | HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and archaea [J]. Science, 2010, 327(5962): 167-170. |
| [7] | CHEN K, WANG Y, ZHANG R, et al.. CRISPR/cas genome editing and precision plant breeding in agriculture [J]. Annu. Rev. Plant Biol., 2019, 70: 667-697. |
| [8] | CHEN J, WANG H, BAI J, et al.. Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163 [J]. Int. J. Biol. Sci., 2019, 15(2): 481-492. |
| [9] | RAN F A, HSU P D, WRIGHT J, et al.. Genome engineering using the CRISPR-Cas9 system [J]. Nat. Protoc., 2013, 8(11): 2281-2308. |
| [10] | FAN Z, LIU Z, XU K, et al.. Long-term, multidomain analyses to identify the breed and allelic effects in MSTN-edited pigs to overcome lameness and sustainably improve nutritional meat production [J]. Sci. China Life Sci., 2022, 65(2): 362-375. |
| [11] | CHEN J, WANG H, BAI J, et al.. Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163 [J]. Int. J. Biol. Sci., 2019, 15(2): 481-492. |
| [12] | LUO L, WANG S, ZHU L, et al.. Aminopeptidase N-null neonatal piglets are protected from transmissible gastroenteritis virus but not porcine epidemic diarrhea virus [J/OL]. Sci. Rep., 2019, 9(1): 13186 [2025-03-20]. . |
| [13] | KABADI A M, OUSTEROUT D G, HILTON I B, et al.. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector [J/OL]. Nucleic Acids Res., 2014, 42(19): e147 [2025-03-20]. . |
| [14] | DONG F, XIE K, CHEN Y, et al.. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells [J]. Biochem. Biophys. Res. Commun., 2017, 482(4): 889-895. |
| [15] | ZHANG J Q, GUO J X, WU X J, et al.. Optimization of sgRNA expression strategy to generate multiplex gene-edited pigs [J]. Zool. Res., 2022, 43(6): 1005-1008. |
| [16] | HAURWITZ R E, STERNBERG S H, DOUDNA J A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA [J]. EMBO J., 2012, 31(12): 2824-2832. |
| [17] | KISHIMOTO T, NISHIMURA K, MORISHITA K, et al.. An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors [J/OL]. J. Biol. Eng., 2024, 18(1): 9 [2025-03-20]. . |
| [18] | GAO Y, ZHAO Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing [J]. J. Integr. Plant Biol., 2014, 56(4): 343-349. |
| [19] | WANG H, SHEN L, CHEN J, et al.. Deletion of CD163 exon 7 confers resistance to highly pathogenic porcine reproductive and respiratory viruses on pigs [J]. Int. J. Biol. Sci., 2019, 15(9): 1993-2005. |
| [20] | CALVERT J G, SLADE D E, SHIELDS S L, et al.. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses [J]. J. Virol., 2007, 81(14): 7371-7379. |
| [21] | PATTON J B, ROWLAND R R, YOO D, et al.. Modulation of CD163 receptor expression and replication of porcine reproductive and respiratory syndrome virus in porcine macrophages [J]. Virus Res., 2009, 140(1-2): 161-171. |
| [22] | VAN GORP H, VAN BREEDAM W, VAN DOORSSELAERE J, et al.. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus [J]. J. Virol., 2010, 84(6): 3101-3105. |
| [23] | JI C M, WANG B, ZHOU J, et al.. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into vero or porcine small intestine epithelial cells [J]. Virology, 2018, 517: 16-23. |
| [24] | ZHU X, LIU S, WANG X, et al.. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection [J/OL]. Emerg. Microbes Infect., 2018, 7(1): 65 [2025-03-20]. . |
| [25] | WHITWORTH K M, ROWLAND R R R, PETROVAN V, et al.. Resistance to coronavirus infection in amino peptidase N-deficient pigs [J]. Transgenic Res., 2019, 28(1): 21-32. |
| [26] | PENG D W, LI R Q, ZENG W, et al.. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Yi Chuan, 2021, 43(3): 261-270. |
| [27] | YANG Z, VAJTA G, XU Y, et al.. Production of pigs by hand-made cloning using mesenchymal stem cells and fibroblasts [J]. Cell. Reprogram., 2016, 18(4): 256-263. |
| [28] | YOSHIOKA K, SUZUKI C, TANAKA A, et al.. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium [J]. Biol. Reprod., 2002, 66(1): 112-119. |
| [29] | 晏超, 刘永刚, 谢浩, 等. 预扩增qPCR技术检测少量猪早期胚胎细胞基因表达的研究[J]. 畜牧兽医学报, 2024, 55(12):5567-5574. |
| YAN C, LIU Y G, XIE H, et al.. Detection of gene expression in trace cells of early porcine embryo by pre-amplified quantitative PCR [J]. Acta Vet. Zootech. Sin., 2024, 55(12): 5567-5574. | |
| [30] | 刘雯雯, 董发明, 毕延震. 多基因编辑技术的发展及其在畜牧种质创新中的应用[J]. 畜牧兽医学报, 2024, 55(8): 3267-3275. |
| LIU W W, DONG F M, BI Y Z. The development of multi-gene editing technology and its application in agricultural biological germplasm innovation [J]. Acta Vet. Zootech. Sin., 2024, 55(8): 3267-3275. | |
| [31] | 郎楠, 梁洛瑜, 汪军丽, 等. CRISPR-Cas9多基因编辑技术在植物研究中的应用[J]. 分子植物育种, 2023, 21(8): 2665-2670. |
| LANG N, LIANG L Y, WANG J L, et al.. Application of CRISPR-Cas9 enabled multiplex gene editing in plant research [J]. Mol. Plant Breeding, 2023, 21(8): 2665-2670. | |
| [32] | XU K, ZHOU Y, MU Y, et al.. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance [J/OL]. eLife, 2020, 9: e57132 [2025-03-20]. . |
| [33] | SAKUMA T, NISHIKAWA A, KUME S, et al.. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system [J/OL]. Sci. Rep., 2014, 4: 5400 [2025-03-20]. . |
| [34] | VAD-NIELSEN J, LIN L, BOLUND L, et al.. Golden gate assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes [J]. Cell. Mol. Life Sci., 2016, 73(22): 4315-4325. |
| [35] | MA X, ZHANG Q, ZHU Q, et al.. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants [J]. Mol. Plant, 2015, 8(8): 1274-1284. |
| [36] | SONG R, WANG Y, ZHENG Q, et al.. One-step base editing in multiple genes by direct embryo injection for pig trait improvement [J]. Sci. China Life Sci., 2022, 65(4): 739-752. |
| [37] | REN J, HAI T, CHEN Y, et al.. Improve meat production and virus resistance by simultaneously editing multiple genes in livestock using Cas12i (Max) [J]. Sci. China Life Sci., 2024, 67(3): 555-564. |
| [38] | ETZERODT A, KJOLBY M, NIELSEN M J, et al.. Plasma clearance of hemoglobin and haptoglobin in mice and effect of CD163 gene targeting disruption [J]. Antioxid. Redox Signal., 2013, 18(17): 2254-2263. |
| [39] | ZHANG Z, BAXTER A E, REN D, et al.. Efficient engineering of human and mouse primary cells using peptide-assisted genome editing [J]. Nat. Biotechnol., 2024, 42(2): 305-315. |
| [40] | XIE K, MINKENBERG B, YANG Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system [J]. Proc. Natl. Acad. Sci. USA, 2015, 112(11): 3570-3575. |
| [41] | ZHANG D, ZHANG H, LI T, et al.. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases [J/OL]. Genome Biol., 2017, 18(1): 191 [2025-03-20]. . |
| [42] | KONSTANTAKOS V, NENTIDIS A, KRITHARA A, et al.. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning [J]. Nucleic Acids Res., 2022, 50(7): 3616-3637. |
| [43] | XU X, DUAN D, CHEN S J. CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment [J/OL]. Sci. Rep., 2017, 7(1): 143 [2025-03-20]. . |
| [44] | DOENCH J G, FUSI N, SULLENDER M, et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 [J]. Nat. Biotechnol., 2016, 34(2): 184-191. |
| [45] | HAN H A, PANG J K S, B-SSOH. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing [J]. J. Mol. Med. (Berl), 2020, 98(5): 615-632. |
| [46] | GUO C, MA X, GAO F, et al.. Off-target effects in CRISPR/Cas9 gene editing [J/OL]. Front. Bioeng. Biotechnol., 2023, 11: 1143157 [2025-03-20]. . |
| [47] | SCHMID-BURGK J L, GAO L, LI D, et al.. Highly parallel profiling of Cas9 variant specificity [J]. Mol. Cell, 2020, 78(4): 794-800.e8. |
| [48] | SCHUSTERBAUER V, FISCHER J E, GANGL S, et al.. Whole genome sequencing analysis of effects of CRISPR/Cas9 in Komagataella phaffii: a budding yeast in distress [J/OL]. J. Fungi (Basel), 2022, 8(10): 992 [2025-03-20]. . |
| [49] | H-HTSAI, H-JKAO, KUO M W, et al.. Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing [J/OL]. Nat. Commun., 2023, 14(1): 5183 [2025-03-20]. . |
| [1] | Wenting ZHANG, Yang LI, Shi QIU, Guangming LU, Dongshu GUO, Baolong ZHANG, Jinyan WANG. Effects of Badh2 Gene on Rice Quality Based on CRISPR/Cas9 Gene Editing Technology [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 39-48. |
| [2] | Caiyong ZHANG, Zhilong CHEN, Hao XIE, Cuiting PENG, Chao YAN, Yulan ZHAO, Lin QI, Lei ZHOU, Zhonglin TANG. Progress of the Improvement of Development of Porcine Somatic Cell Nuclear Transfer Embryos [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 201-209. |
| [3] | Zixin LI, Hongfei BAI, Yong XIE, Xun LI, Lijing BAI. Establishment of CSE1L Knockout C2C12 Cells by CRISPR-Cas9 System [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 225-233. |
| [4] | Qiangzhou WANG, Shiyu PAN, Mengya FANG, Wei LI, Jiaxing WANG, Yinyin LIU, Shihao CHEN. Construction of Chicken Embryo Fibroblast Cell Line with TET2 Gene Knockout Based on CRISPR-Cas9 Technology [J]. Journal of Agricultural Science and Technology, 2023, 25(11): 227-233. |
| [5] | Libin WANG, Qianglong WANG, Yangyang PAN, Tian ZHAO, Tianyi DING, Yan CUI, Sijiu YU. In Vitro Maturation Culture and Parthenogenesis of Yak Oocytes [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 84-90. |
| [6] | Chundi XIE, Xiaorong GUO, Xudong ZHANG, Rong ZHOU, Kui LI. Establishment of IGF2R Knockout PK-15 Cells by CRISPR/Cas9 System [J]. Journal of Agricultural Science and Technology, 2022, 24(10): 200-207. |
| [7] | Zhong ZHUANG, Long ZHAO, Hao BAI, Yulin BI, Yingquan HUANG, Guohong CHEN, Guobin CHANG. Development of CRISPR/Cas9 and Its Application in Poultry [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 14-23. |
| [8] | BAIGETUMUER Ayitula, LI jie, CHEN Cunkun, LIU hui, LI Xiangyang, LIN Shaohua. Combined Effect of Grapefruit Seed Extract Treatment and Polyethylene Packaging on Postharvest Quality of Toona Sinensis [J]. Journal of Agricultural Science and Technology, 2021, 23(5): 116-123. |
| [9] | LIAO Jiaming, §, LI Chunmei§, ZHANG Shihu, LI Buye, OUYANG Kunxi, CHEN Xiaoyang. Development of CRISPR/Cas9 System and Its Aapplication in Plants [J]. Journal of Agricultural Science and Technology, 2021, 23(12): 20-28. |
| [10] | XIN Hongjia, LI Pengcheng, TENG Shouzhen, LI Shengyan, WANG Hai, LANG Zhihong*. Construction and Functional Characterization of Mutants of Arabidopsis SWEET1/2/3 Genes [J]. Journal of Agricultural Science and Technology, 2020, 22(2): 39-49. |
| [11] | LIU Dujuan, HUANG Huoqing*, SU Xiaoyun*. Construction of a Trichoderma reesei Strain with Low Cellulase Background and Its Application [J]. Journal of Agricultural Science and Technology, 2020, 22(12): 50-57. |
| [12] | HU Wan-bin, LI Jia-xiang, DUAN Li-zhu, LIU Min-bo, CHANG Ya-qing, . The Impact of Seawater Acidification on Early Development of the Sea Urchin Hemicentrotus pulcherrimus [J]. Journal of Agricultural Science and Technology, 2016, 18(3): 177-183. |
| [13] | WANG Cheng-long1,2, ZHOU Mei-liang2, DONG Xue-ni1,2, TANG Yi-xiong2, SHAO Ji-ron. Optimization and Comparison of Two Regeneration System of Alfalfa (Medicago sativa L.) [J]. , 2015, 17(4): 53-61. |
| [14] | LI Xi-he1,2 . Research and Development of Livestock Reproductive Biotechnology and its Industrialization and Popularization [J]. , 2013, 15(3): 64-71. |
| [15] | Hunter R H F1, LI Xihe2,3*. EggEmbryo Transfer: An Analytical Tool for Vintage Experiments in Domestic Farm Animals [J]. , 2013, 15(1): 65-70. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号