




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (2): 27-37.DOI: 10.13304/j.nykjdb.2022.0612
梁婷1( ), 左静红2, 陆青1, 杨东2, 唐益苗2, 郭春曼2(
), 左静红2, 陆青1, 杨东2, 唐益苗2, 郭春曼2( ), 汪德州2(
), 汪德州2( ), 王伟伟2(
), 王伟伟2( )
)
收稿日期:2022-07-19
接受日期:2022-09-28
出版日期:2023-02-15
发布日期:2023-05-17
通讯作者:
郭春曼,汪德州,王伟伟
作者简介:梁婷 E-mail:1577033136@qq.com
基金资助:
Ting LIANG1( ), Jinghong ZUO2, Qing LU1, Dong YANG2, Yimiao TANG2, Chunman GUO2(
), Jinghong ZUO2, Qing LU1, Dong YANG2, Yimiao TANG2, Chunman GUO2( ), Dezhou WANG2(
), Dezhou WANG2( ), Weiwei WANG2(
), Weiwei WANG2( )
)
Received:2022-07-19
Accepted:2022-09-28
Online:2023-02-15
Published:2023-05-17
Contact:
Chunman GUO,Dezhou WANG,Weiwei WANG
摘要:
IQM基因是钙调素结合蛋白家族中的重要分支,在植物生长发育和应激反应中发挥重要作用。本研究利用生物信息学方法在小麦全基因组中鉴定出23个IQM基因家族成员,对其染色体位置、理化性质、系统进化关系、基因结构、蛋白保守结构域、启动子顺式作用元件和基因表达特性等进行了系统分析。结果表明,TaIQM基因家族成员随机分布在小麦18条染色体上,亚细胞定位结果显示所有基因均位于细胞核中,系统进化分析将其分为3个亚类,同一亚类间基因结构、蛋白保守结构域相似度较高,推测其功能相似;顺式作用元件分析表明,TaIQM基因家族成员启动子中含有多种与逆境胁迫及生长发育相关顺式作用元件;转录组数据分析显示,TaIQM基因家族成员在不同时期的根、茎、叶、穗和籽粒中表达量存在显著性差异;qRT-PCR分析显示,在小麦苗期地上部和地下部,TaIQM基因响应干旱、低温、高温、NaCl、ABA等多种胁迫,显示上调或下调表达。初步推断这些基因可能通过Ca2+信号通路参与非生物胁迫调控,研究结果为全面解析TaIQM基因结构与生物学功能、非生物胁迫响应分子机制提供依据。
中图分类号:
梁婷, 左静红, 陆青, 杨东, 唐益苗, 郭春曼, 汪德州, 王伟伟. 小麦IQM基因家族鉴定及非生物胁迫下表达分析[J]. 中国农业科技导报, 2023, 25(2): 27-37.
Ting LIANG, Jinghong ZUO, Qing LU, Dong YANG, Yimiao TANG, Chunman GUO, Dezhou WANG, Weiwei WANG. Identification and Expression Analysis Under Abiotic Stress of IQM Gene Family in Wheat (Triticum aestivum L.)[J]. Journal of Agricultural Science and Technology, 2023, 25(2): 27-37.
| 基因 Gene | 正向引物 Forward primer (5’–3’) | 反向引物 Reverse primer (5’–3’) |
|---|---|---|
| TaIQM1 | GACGATTGGTGGTCAAAGATGG | ACGCCGTCTATCTCAACTTCTGTC |
| TaIQM2 | TCGTTTCAGCACTCCAGTTTCC | TTGACATCGGTGAGGCTAACGT |
| TaIQM3 | CAAGGGCAGTGCGTCAAGTA | TGGAGTCATCCGAAGTGTGC |
| TaIQM4 | GGAGATGTTCACAAGGCTGGATTC | GGGTAGTCCCTGACGCAAATGAT |
| TaIQM5 | ATGGAGTTCTGAAGGCTATCTGGC | TTCTTCGGGTTTGTCGGCATC |
| TaIQM6 | GTGCCAAGGGAGAAGGTCATT | GGTAGTCCCTGACGCAAATGAT |
| TaIQM7 | CGAGCCTACTGAAACGGAAGAAC | CCATTTGAAGGACGGGAGACTG |
| TaIQM8 | TCAAGAACTGGGAGGCGGAG | AGACGAAGTGCGGCTTGGAC |
| TaIQM9 | CGAGAGGGCAAGGTATGAGGTTA | ACCCTTCTTCTTCTGCCCGAT |
| TaIQM10 | AACTCGGACAGGCCATTTGT | TAGAGCTCACCGCATCGAAC |
| TaIQM11 | CCATAAGCACAACCACAGCCT | CAACAATGGACGACAAGCACAG |
| TaIQM12 | CTCGCCAGCAGATGATGACA | AAGGACCCTTTTCTCGCAGG |
| TaIQM13 | GCTCATCCACCAAGGAAGACTAC | GGTACCATGTTGGAAGGCAGT |
| TaIQM14 | GGAACCCTGAAGGCTATTTGG | TGGTTCCTCCCGTTTCTCTGT |
| TaIQM15 | TAATGTCCGTGTGGAGCAAGCA | CGCTTGCATTCTGAGCTCTATCG |
| TaIQM16 | GTGGTCACTACCGCCCTACA | CCTCTGCTGGGCTCATCTTC |
| TaIQM17 | TGGAGCAAGCGAGACCAACCTA | GGCCGAGCTGATAGGATTTTGAC |
| TaIQM18 | GAGAGGACTATGAGGTCGTGATTG | CCATTTTCAGCAACCAGCCT |
| TaIQM19 | CCCAACTGATTCTCCCTTCCAAC | CCACACGGAGATTATCGTTGACCT |
| TaIQM20 | CACAACTGATTCTCCCTTCCAAC | ATTCCCAGTGCTCCATTTCAG |
| TaIQM21 | AGTGGACATTACAAACCAAGTGCG | ATTGTGGAGGATTGGATGGCA |
| TaIQM22 | TGGACATTACAAACCGAGTGCG | AGCAGCAGTAGGGTTCTGTTTGGT |
| TaIQM23 | CGTCAGCCTCAACGCATCAA | GGTTATGCCCGTACCTATGTCTTG |
| 18S | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
表1 qRT-PCR引物 (续表Continued)
Table 1 qRT-PCR primers
| 基因 Gene | 正向引物 Forward primer (5’–3’) | 反向引物 Reverse primer (5’–3’) |
|---|---|---|
| TaIQM1 | GACGATTGGTGGTCAAAGATGG | ACGCCGTCTATCTCAACTTCTGTC |
| TaIQM2 | TCGTTTCAGCACTCCAGTTTCC | TTGACATCGGTGAGGCTAACGT |
| TaIQM3 | CAAGGGCAGTGCGTCAAGTA | TGGAGTCATCCGAAGTGTGC |
| TaIQM4 | GGAGATGTTCACAAGGCTGGATTC | GGGTAGTCCCTGACGCAAATGAT |
| TaIQM5 | ATGGAGTTCTGAAGGCTATCTGGC | TTCTTCGGGTTTGTCGGCATC |
| TaIQM6 | GTGCCAAGGGAGAAGGTCATT | GGTAGTCCCTGACGCAAATGAT |
| TaIQM7 | CGAGCCTACTGAAACGGAAGAAC | CCATTTGAAGGACGGGAGACTG |
| TaIQM8 | TCAAGAACTGGGAGGCGGAG | AGACGAAGTGCGGCTTGGAC |
| TaIQM9 | CGAGAGGGCAAGGTATGAGGTTA | ACCCTTCTTCTTCTGCCCGAT |
| TaIQM10 | AACTCGGACAGGCCATTTGT | TAGAGCTCACCGCATCGAAC |
| TaIQM11 | CCATAAGCACAACCACAGCCT | CAACAATGGACGACAAGCACAG |
| TaIQM12 | CTCGCCAGCAGATGATGACA | AAGGACCCTTTTCTCGCAGG |
| TaIQM13 | GCTCATCCACCAAGGAAGACTAC | GGTACCATGTTGGAAGGCAGT |
| TaIQM14 | GGAACCCTGAAGGCTATTTGG | TGGTTCCTCCCGTTTCTCTGT |
| TaIQM15 | TAATGTCCGTGTGGAGCAAGCA | CGCTTGCATTCTGAGCTCTATCG |
| TaIQM16 | GTGGTCACTACCGCCCTACA | CCTCTGCTGGGCTCATCTTC |
| TaIQM17 | TGGAGCAAGCGAGACCAACCTA | GGCCGAGCTGATAGGATTTTGAC |
| TaIQM18 | GAGAGGACTATGAGGTCGTGATTG | CCATTTTCAGCAACCAGCCT |
| TaIQM19 | CCCAACTGATTCTCCCTTCCAAC | CCACACGGAGATTATCGTTGACCT |
| TaIQM20 | CACAACTGATTCTCCCTTCCAAC | ATTCCCAGTGCTCCATTTCAG |
| TaIQM21 | AGTGGACATTACAAACCAAGTGCG | ATTGTGGAGGATTGGATGGCA |
| TaIQM22 | TGGACATTACAAACCGAGTGCG | AGCAGCAGTAGGGTTCTGTTTGGT |
| TaIQM23 | CGTCAGCCTCAACGCATCAA | GGTTATGCCCGTACCTATGTCTTG |
| 18S | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
基因 Gene | 基因号 Gene ID | 染色体定位 Chromosome localization | 氨基酸数 Number of amino acids | 相对分子量 Molecular weight/Da | 等电点 Point isoelectric | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TaIQM1 | TraesCS1A02G125400.1 | Chr1A: 149352287-149356056 | 517 | 57 811.59 | 7.68 | 细胞核Nucleus |
| TaIQM2 | TraesCS1B02G143800.1 | Chr1B: 195555337-195560543 | 580 | 65 214.18 | 7.69 | 细胞核 Nucleus |
| TaIQM3 | TraesCS1D02G128400.1 | Chr1D: 141545585-141547817 | 424 | 47 667.67 | 9.49 | 细胞核Nucleus |
| TaIQM4 | TraesCS2A02G154900.1 | Chr2A: 102130189-102132879 | 564 | 63 624.94 | 6.47 | 细胞核Nucleus |
| TaIQM5 | TraesCS2B02G180000.1 | Chr2B: 154592307-154595019 | 572 | 64 351.52 | 6.36 | 细胞核Nucleus |
| TaIQM6 | TraesCS2D02G160200.1 | Chr2D: 103126893-103129599 | 571 | 64 331.63 | 6.26 | 细胞核Nucleus |
| TaIQM7 | TraesCS3A02G206400.1 | Chr3A: 363415357-363419189 | 556 | 62 188.59 | 9.13 | 细胞核Nucleus |
| TaIQM8 | TraesCS3B02G238500.1 | Chr3B: 373842628-373844908 | 541 | 60 758.12 | 9.07 | 细胞核Nucleus |
| TaIQM9 | TraesCS3D02G209200.1 | Chr3D: 276574646-276577885 | 530 | 59 496.51 | 8.97 | 细胞核Nucleus |
| TaIQM10 | TraesCS4A02G021000.1 | Chr4A: 14261744-14265762 | 610 | 67 879.37 | 9.60 | 细胞核Nucleus |
| TaIQM11 | TraesCS4B02G282600.2 | Chr4B: 565861371-565865127 | 669 | 74 779.20 | 9.41 | 细胞核Nucleus |
| TaIQM12 | TraesCS4D02G281600.1 | Chr4D: 452795390-452798654 | 613 | 68 128.53 | 9.67 | 细胞核Nucleus |
| TaIQM13 | TraesCS5A02G129600.1 | Chr5A: 290924878-290930090 | 461 | 51 276.55 | 6.88 | 细胞核Nucleus |
| TaIQM14 | TraesCS5A02G375700.1 | Chr5A: 573606336-573609441 | 534 | 59 693.35 | 8.60 | 细胞核Nucleus |
| TaIQM15 | TraesCS5B02G128200.1 | Chr5B: 234295411-234304552 | 480 | 53 236.89 | 6.14 | 细胞核Nucleus |
| TaIQM16 | TraesCS5B02G377900.1 | Chr5B: 555950716-555953748 | 538 | 60 186.87 | 7.57 | 细胞核Nucleus |
| TaIQM17 | TraesCS5D02G137100.1 | Chr5D: 218051693-218055671 | 480 | 53 123.76 | 6.30 | 细胞核Nucleus |
| TaIQM18 | TraesCS5D02G385200.1 | Chr5D: 454309041-454312175 | 533 | 59 832.36 | 7.17 | 细胞核Nucleus |
| TaIQM19 | TraesCS6A02G108900.1 | Chr6A: 77780102-77784144 | 485 | 53 610.33 | 8.58 | 细胞核Nucleus |
| TaIQM20 | TraesCS6B02G133900.1 | Chr6B: 130822739-130829829 | 477 | 52 647.25 | 6.44 | 细胞核Nucleus |
| TaIQM21 | TraesCS6B02G137500.1 | Chr6B: 135112248-135116198 | 483 | 53 349.14 | 8.82 | 细胞核Nucleus |
| TaIQM22 | TraesCS6D02G093400.1 | Chr6D: 58234159-58240593 | 480 | 53 062.87 | 8.04 | 细胞核Nucleus |
| TaIQM23 | TraesCS6D02G097300.1 | Chr6D: 61134482-61138500 | 484 | 53 425.06 | 8.06 | 细胞核Nucleus |
表2 小麦IQM基因家族蛋白理化性质及亚细胞定位 (续表Continued)
Table 2 Characteristics and subcellular localization prediction of TaIQM genes
基因 Gene | 基因号 Gene ID | 染色体定位 Chromosome localization | 氨基酸数 Number of amino acids | 相对分子量 Molecular weight/Da | 等电点 Point isoelectric | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TaIQM1 | TraesCS1A02G125400.1 | Chr1A: 149352287-149356056 | 517 | 57 811.59 | 7.68 | 细胞核Nucleus |
| TaIQM2 | TraesCS1B02G143800.1 | Chr1B: 195555337-195560543 | 580 | 65 214.18 | 7.69 | 细胞核 Nucleus |
| TaIQM3 | TraesCS1D02G128400.1 | Chr1D: 141545585-141547817 | 424 | 47 667.67 | 9.49 | 细胞核Nucleus |
| TaIQM4 | TraesCS2A02G154900.1 | Chr2A: 102130189-102132879 | 564 | 63 624.94 | 6.47 | 细胞核Nucleus |
| TaIQM5 | TraesCS2B02G180000.1 | Chr2B: 154592307-154595019 | 572 | 64 351.52 | 6.36 | 细胞核Nucleus |
| TaIQM6 | TraesCS2D02G160200.1 | Chr2D: 103126893-103129599 | 571 | 64 331.63 | 6.26 | 细胞核Nucleus |
| TaIQM7 | TraesCS3A02G206400.1 | Chr3A: 363415357-363419189 | 556 | 62 188.59 | 9.13 | 细胞核Nucleus |
| TaIQM8 | TraesCS3B02G238500.1 | Chr3B: 373842628-373844908 | 541 | 60 758.12 | 9.07 | 细胞核Nucleus |
| TaIQM9 | TraesCS3D02G209200.1 | Chr3D: 276574646-276577885 | 530 | 59 496.51 | 8.97 | 细胞核Nucleus |
| TaIQM10 | TraesCS4A02G021000.1 | Chr4A: 14261744-14265762 | 610 | 67 879.37 | 9.60 | 细胞核Nucleus |
| TaIQM11 | TraesCS4B02G282600.2 | Chr4B: 565861371-565865127 | 669 | 74 779.20 | 9.41 | 细胞核Nucleus |
| TaIQM12 | TraesCS4D02G281600.1 | Chr4D: 452795390-452798654 | 613 | 68 128.53 | 9.67 | 细胞核Nucleus |
| TaIQM13 | TraesCS5A02G129600.1 | Chr5A: 290924878-290930090 | 461 | 51 276.55 | 6.88 | 细胞核Nucleus |
| TaIQM14 | TraesCS5A02G375700.1 | Chr5A: 573606336-573609441 | 534 | 59 693.35 | 8.60 | 细胞核Nucleus |
| TaIQM15 | TraesCS5B02G128200.1 | Chr5B: 234295411-234304552 | 480 | 53 236.89 | 6.14 | 细胞核Nucleus |
| TaIQM16 | TraesCS5B02G377900.1 | Chr5B: 555950716-555953748 | 538 | 60 186.87 | 7.57 | 细胞核Nucleus |
| TaIQM17 | TraesCS5D02G137100.1 | Chr5D: 218051693-218055671 | 480 | 53 123.76 | 6.30 | 细胞核Nucleus |
| TaIQM18 | TraesCS5D02G385200.1 | Chr5D: 454309041-454312175 | 533 | 59 832.36 | 7.17 | 细胞核Nucleus |
| TaIQM19 | TraesCS6A02G108900.1 | Chr6A: 77780102-77784144 | 485 | 53 610.33 | 8.58 | 细胞核Nucleus |
| TaIQM20 | TraesCS6B02G133900.1 | Chr6B: 130822739-130829829 | 477 | 52 647.25 | 6.44 | 细胞核Nucleus |
| TaIQM21 | TraesCS6B02G137500.1 | Chr6B: 135112248-135116198 | 483 | 53 349.14 | 8.82 | 细胞核Nucleus |
| TaIQM22 | TraesCS6D02G093400.1 | Chr6D: 58234159-58240593 | 480 | 53 062.87 | 8.04 | 细胞核Nucleus |
| TaIQM23 | TraesCS6D02G097300.1 | Chr6D: 61134482-61138500 | 484 | 53 425.06 | 8.06 | 细胞核Nucleus |
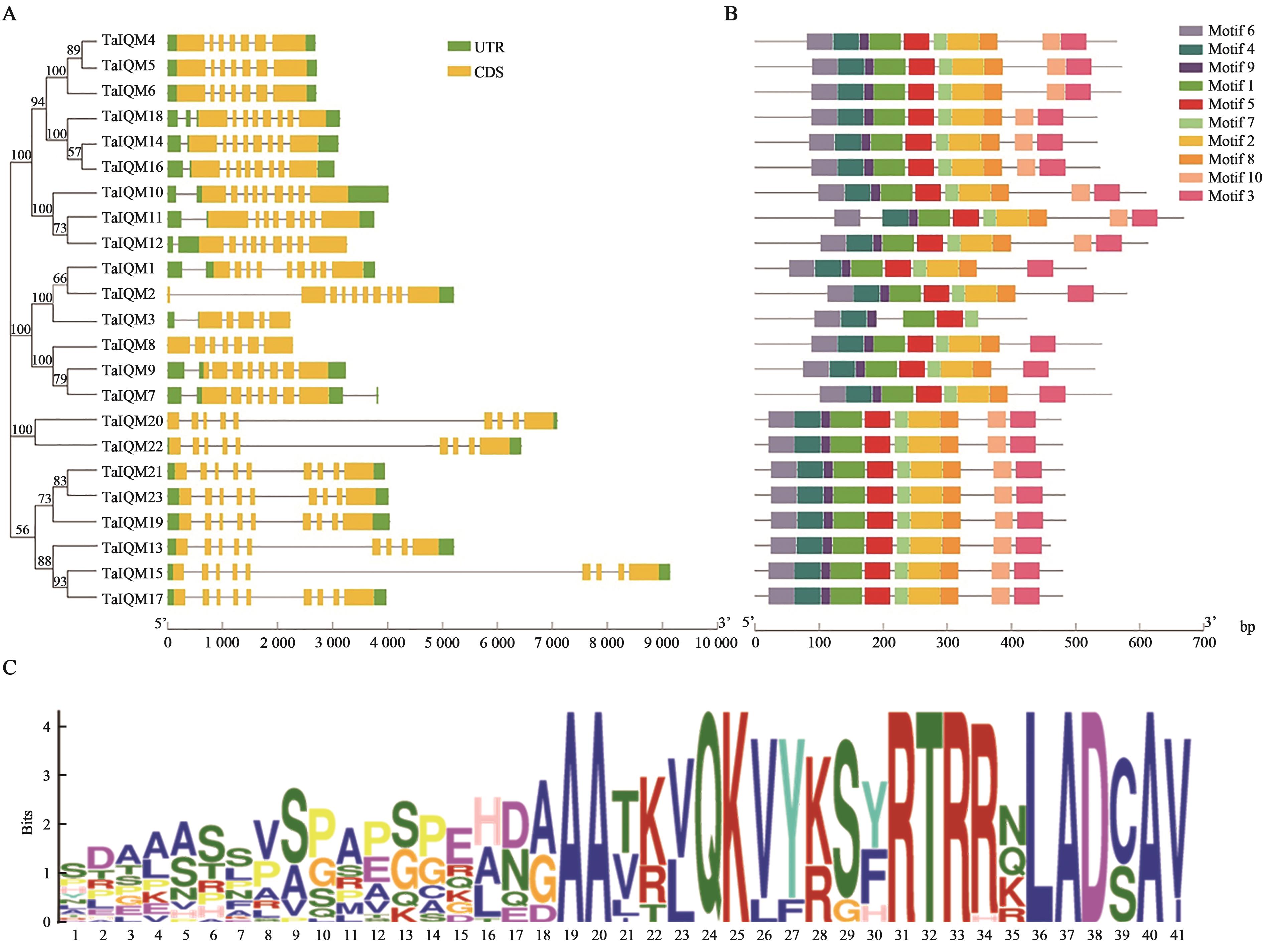
图2 TaIQM家族成员结构特征和保守基序分析A:基因结构;B:保守基序;C:IQM保守基序序列
Fig. 2 Structure and conservative motif analysis of the TaIQMA: Gene structure;B: Conserved motif;C: Conserved motif sequences of IQM

图3 TaIQM启动子顺式作用元件预测注:ABRE—脱落酸顺式作用元件;ARE—无氧诱导所必需的顺式作用调节元件;Circadian—参与昼夜节律控制的顺式作用调节元件;G-box—参与光反应的顺式作用调控元件;TGACG-motif—与 MeJA 反应有关的顺式作用调节因子;GT1-motif—光响应元件;TC-rich repeats—参与防御和应激反应的顺式作用元件;G-box—参与光反应的顺式作用调控元件;CGTCA-motif—缺氧特异性诱导中的增强子样元件;CAT-box—与分生组织表达有关的顺式作用调控元件;P-box—赤霉素反应元件;GARE-motif—赤霉素反应元件;TGA-element—生长素反应元件;ACE—参与光反应的顺式作用元件;GCN4_motif—参与胚乳表达顺式作用元件;TCA-element—参与水杨酸反应的顺式作用元件;TATC-box—赤霉素反应顺式作用元件;MBS—干旱诱导MYB结合位点;AuxRR-core—参与生长素反应的顺式作用元件;LTR—参与低温响应的顺式作用元件。
Fig. 3 Cis-acting elements analysis of TaIQM genesNote: ABRE—Cis-acting element involved in the abscisic acid responsiveness; ARE—Cis-acting regulatory element essential for the anaerobic induction; Circadian—Cis-acting regulatory element involved in circadian control; G-box—Cis-acting regulatory element involved in light responsiveness; TGACG-motif—Cis-acting regulatory element involved in the MeJA-responsiveness; GT1-motif—Light responsive element; TC-rich repeats—Cis-acting element involved in defense and stress responsiveness; G-box—Cis-acting regulatory element involved in light responsiveness; CGTCA-motif—Cis-acting regulatory element involved in the MeJA-responsiveness; CAT-box—Cis-acting regulatory element related to meristem expression; P-box—Gibberellin-responsive element; GARE-motif—Gibberellin-responsive element; TGA-element—Auxin-responsive element; ACE—Cis-acting element involved in light responsiveness; GCN4_motif—Cis-regulatory element involved in endosperm expression; TCA-element—Cis-acting element involved in salicylic acid responsiveness; TATC-box—Cis-acting element involved in gibberellin-responsiveness; MBS—MYB binding site involved in drought-inducibility; AuxRR-core—Cis-acting regulatory element involved in auxin responsiveness; LTR—Cis-acting element involved in low-temperature responsiveness.
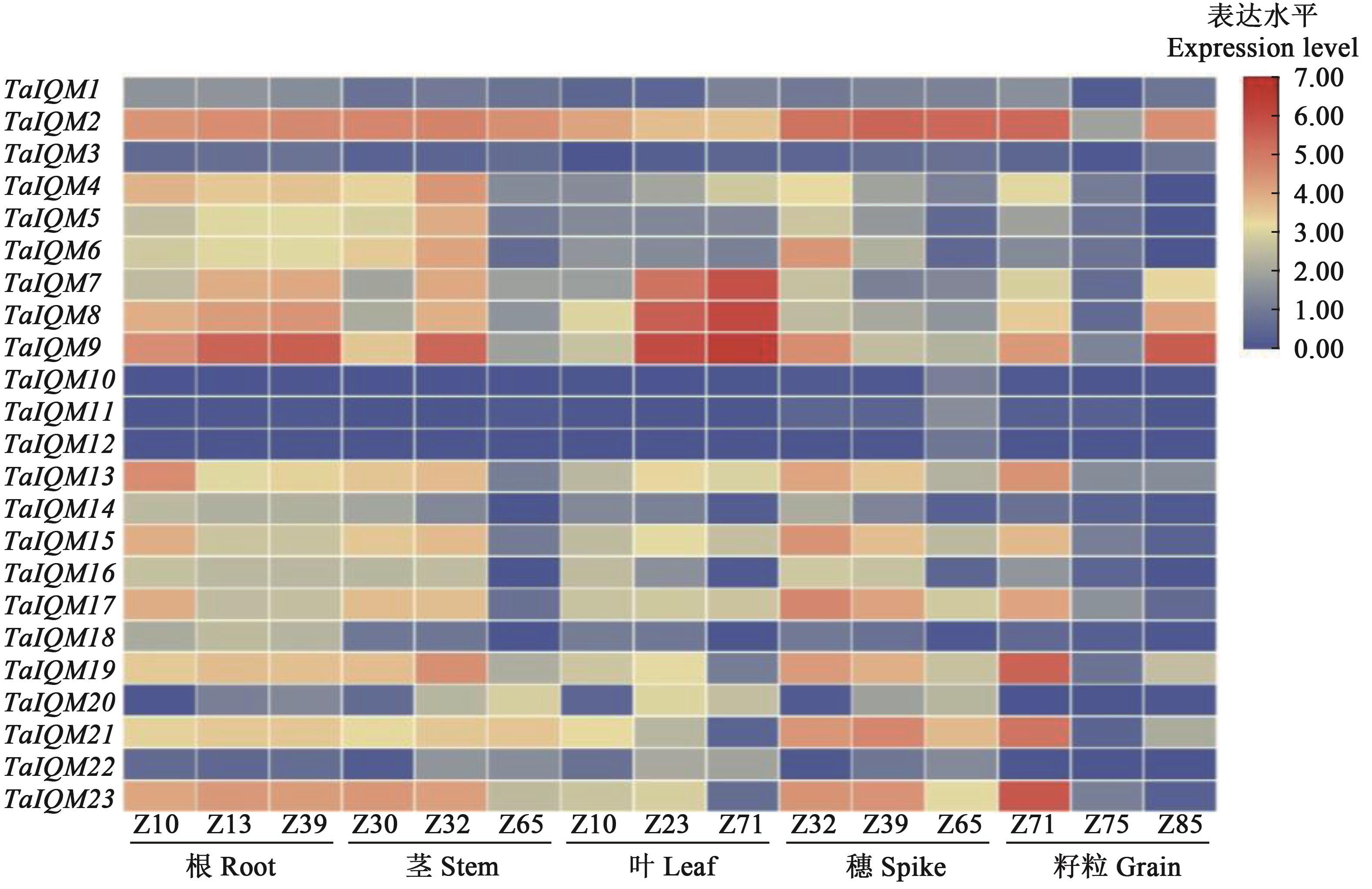
图4 TaIQM基因在不同组织中的表达量热图注:红色表示表达上调,蓝色表示表达下调。Z10—1叶期;Z13—3叶期;Z23—分蘖早期;Z30—起身期;Z32—拔节早期;Z39—拔节晚期;Z65—开花中期;Z71—开花后2 d;Z75—开花后10 d;Z85—开花后30 d。
Fig. 4 Heat map of relative expression level of TaIQM genes in different tissuesNote: Red color indicates up-regulation expression, blue color indicates down-regulation expression. Z10—1 leaf period; Z13—3 leaves stage; Z23—Early tillering; Z30—Standing stage; Z32—Early jointing stage; Z39—Late jointing stage; Z65—Middle flowering; Z71—2 d after flowering; Z75—10 d after flowering; Z85—30 d after flowering.
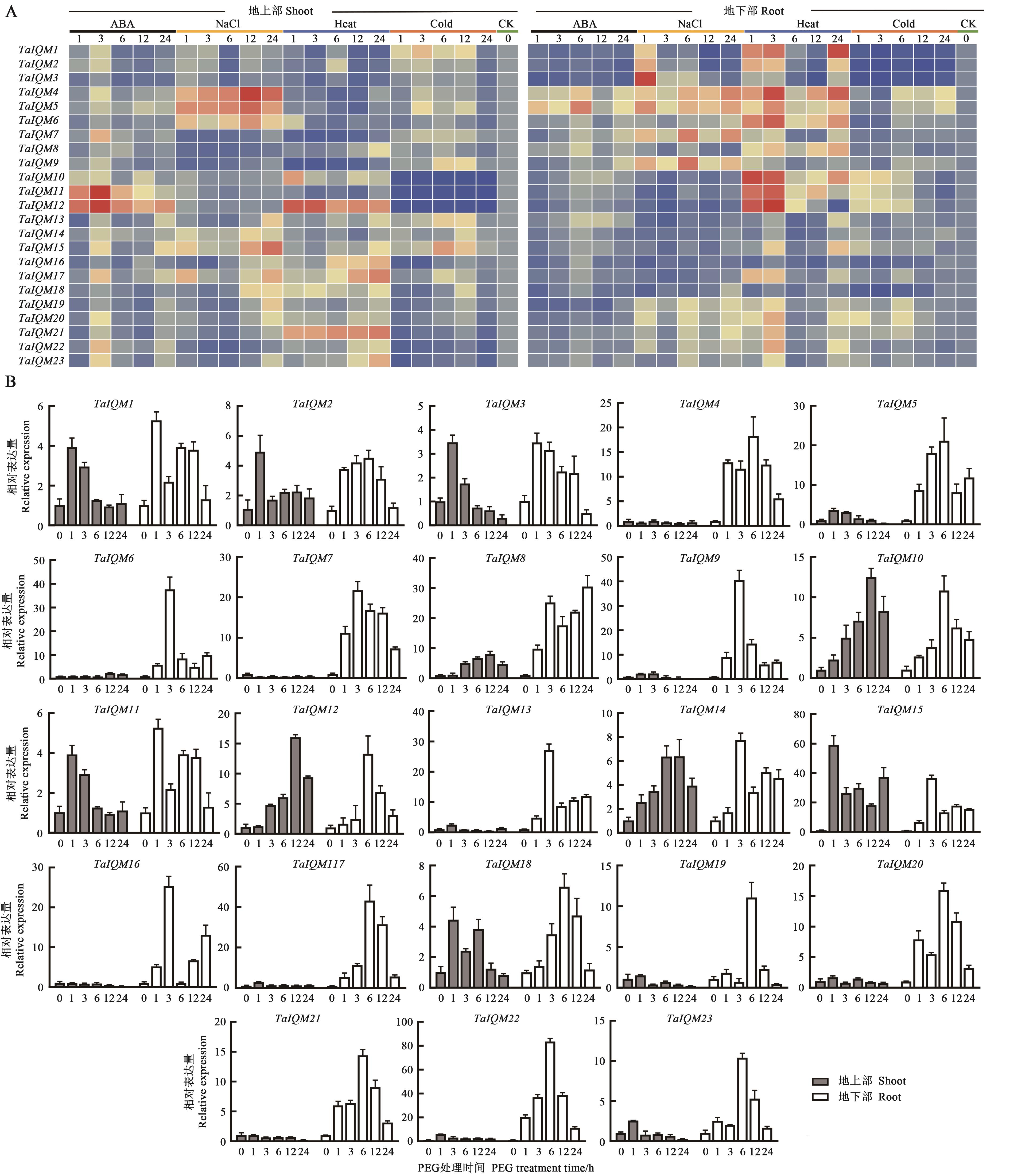
图5 小麦IQM基因家族在非生物胁迫下的表达分析A:ABA、NaCl、热、冷处理表达量分析; B:干旱处理表达量分析。红色表示表达上调,蓝色表示表达下调
Fig. 5 Expression analysis of IQM genes in wheat under abiotic stressesA: Relative expressionunder ABA, NaCl, heat and clod; B: Relative expression under drought.Red color indicates up-regulation expression, blue color indicates down-regulation expression
| 1 | 田长恩, 周玉萍. 植物具IQ基序的钙调素结合蛋白的研究进展[J]. 植物学报, 2013,48(4):447-460. |
| TIAN C N, ZHOU Y P. Research progress in plant IQ motif-containing calmodulin-binding proteins [J]. Chin. Bull. Botany, 2013, 48(4): 447-460. | |
| 2 | ZHOU Y P, DUAN J, FUJIBE T, et al.. AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in Arabidopsis [J]. Plant Mol. Biol., 2012,79(4-5):333-346. |
| 3 | 曾后清, 张亚仙, 汪尚, 等. 植物钙/钙调素介导的信号转导系统[J]. 植物学报, 2016,51(5):705-723. |
| ZENG H Q, ZHANG Y X, WANG S, et al.. Calcium/calmodulin-mediated signal transduction systems in plants [J]. Chin. Bull. Bot., 2016,51(5):705-723. | |
| 4 | DEFALCO T A, BENDER K W, SNEDDEN W A. Breaking the code: Ca2+ sensors in plant signalling [J]. Biochem. J., 2009,425(1):27-40. |
| 5 | NG C K, MCAINSH M R, GRAY J E, et al.. Calcium-based signalling systems in guard cells [J]. New Phytol., 2001,151(1):109-120. |
| 6 | ZHOU Y P, WU J H, XIAO W H, et al.. Arabidopsis IQM 4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in Arabidopsis [J/OL]. Front. Plant Sci., 2018,9:721 [2022-06-08].. |
| 7 | FAN T, LYU T, XIE C, et al.. Genome-wide analysis of the IQM gene family in rice (Oryza sativa L.) [J/OL]. Plants, 2021,10(9):1949 [2022-06-08].. |
| 8 | PATRA N, HARIHARAN S, GAIN H, et al.. Typical but delicate Ca(2+)re: dissecting the essence of calcium signaling network as a robust response coordinator of versatile abiotic and biotic stimuli in plants [J/OL]. Front. Plant Sci., 2021,12:752246 [2022-06-08]. . |
| 9 | 罗慧婷, 吕天晓, 范甜, 等. 拟南芥IQM1互作蛋白的筛选与验证[J]. 科技视界, 2018(9):83-84. |
| LUO H T, LYU T X, FAN T, et al.. Screening and verification of the protein interacting with IQM1 in Arabidopsis [J]. Sci. Technol. Vision, 2018(9):83-84. | |
| 10 | 周玉萍, 陈琼华, 陈洁珊, 等. IQM1基因过量表达对拟南芥气孔运动及根系生长的影响[J]. 西北植物学报, 2013,33(5):904-910. |
| ZHOU Y P, CHEN Q H, CHEN J S, et al.. Overexpression of Arabidopsis IQM1 gene affects stomatal movement and root growth [J]. Acta Bot. Bor-Occid. Sin., 2013,33(5):904-910. | |
| 11 | 黄章科, 张艺能, 莫忠蓁, 等. IQ基序突变对AtIQM1的钙调素结合活性的影响[J]. 生物技术通报, 2012,12(21):128-132. |
| HUANG Z K, ZHANG Y N, MO Z Q, et al.. Effects of mutations in IQ motif of AtIQM1 on its calmodulin binding [J]. Biotechnol. Bull., 2012,12(21):128-132. | |
| 12 | 吴骏. 拟南芥IQM2参与成花调控与CaM信号关系的初步研究[D]. 广州:广州大学, 2017. |
| WU J. Preliminary study on relationship of IQM2 mediating flowering and CaM signaling in Arabidopsis [D]. Guangzhou: Guangzhou University, 2017. | |
| 13 | 徐浩, 冯奕嘉, 范甜, 等. 拟南芥IQM3基因突变减少幼苗的侧根数量和增加主根长度[J]. 植物生理学报, 2019,55(5):629-634. |
| XU H, FENG Y J, FAN T, et al.. Disruption of IQM3 reduces the number of lateral roots and increases the length of primary root in Arabidopsis seedlings [J]. Plant Physiol. J., 2019,55(5):629-634. | |
| 14 | 萧文慧, 宋俊威, 黄小玲, 等. 非生物胁迫对拟南芥IQM4基因表达的影响[J]. 科技视界, 2016(16):10-11. |
| XIAO W H, SONG J W, HUANG X L, et al.. Effect of abiotic stress on IQM4 gene expression in Arabidopsis thaliana [J]. Sci. Technol. Vision, 2016(16):10-11. | |
| 15 | 弓路平, 萧文慧, 周玉萍, 等. 拟南芥IQM5.2的克隆、表达及其生物信息学分析[J]. 生物技术通报, 2016,32(5):69-74. |
| GONG L P, XIAO W H, ZHOU Y P, et al.. Cloning,expression and bioinformatics analysis of IQM5.2 from Arabidopsis [J]. Biotechnol. Bull.,2016,32(5):69-74. | |
| 16 | 冯奕嘉, 徐浩, 范甜, 等. 拟南芥IQM6突变推迟远轴面表皮毛的发生[J]. 植物生理学报, 2019,55(6):729-735. |
| FENG Y J, XU H, FAN T, et al.. IQM6 mutantion delays initiation of abaxial trichomes in Arabidopsis [J]. Plant Physiol. J., 2019,55(6):729-735. | |
| 17 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The pfam protein families database in 2019 [J/OL]. Nucleic Acids Res., 2019,47(D1):995 [2022-06-08]. . |
| 18 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J/OL]. Nucleic Acids Res., 2002,30(1):325 [2022-06-08]. . |
| 19 | AROCHO A, CHEN B, LADANYI M, et al.. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts [J]. Diagn Mol. Pathol., 2006,15(1):56-61. |
| 20 | WU M, LI Y, CHEN D, et al.. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis) [J/OL]. Sci. Rep., 2016,6:24520 [2022-06-08]. . |
| 21 | ZHOU Y, CHEN Y, YAMAMOTO K T, et al.. Sequence and expression analysis of the Arabidopsis IQM family [J]. Acta Physiol. Plantarum., 2010,32(1):191-198. |
| 22 | ZHAO J F, ZHAO L L, ZHANG M, et al.. Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation [J/OL]. Int. J. Mol. Sci., 2017, 18(9): 1841 [2022-06-08]. . |
| 23 | 杨华杰, 周玉萍, 范甜, 等. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021,37(11):190-196. |
| YANG H J, ZHOU Y P, FAN T, et al.. Screening and identification of IQM4-interacting proteins in Arabidopsis thaliana [J]. Biotechnol. Bull., 2021,37(11):190-196. |
| [1] | 刘一凡, 刘少帅, 臧瑞, 李洋, 刘薇, 李婷婷, 刘旦梅, 刘登才, 李爱丽, 毛龙, 王翔, 耿帅锋. 168份小麦种质资源品质性状分析[J]. 中国农业科技导报, 2025, 27(9): 44-57. |
| [2] | 吕彩霞, 李永福, 信会男, 李娜, 赖宁, 耿庆龙, 陈署晃. 缓释氮肥对滴灌冬小麦产量及土壤硝/铵态氮的影响[J]. 中国农业科技导报, 2025, 27(8): 179-186. |
| [3] | 朱强, 车宗贤, 崔恒, 张久东, 包兴国. 绿肥替代氮肥对麦田温室气体的影响[J]. 中国农业科技导报, 2025, 27(7): 182-189. |
| [4] | 胡懿, 公杰, 赵玮, 程蓉, 柳忠玉, 高世庆, 杨亚珍. 小麦PHY基因家族鉴定及热胁迫下表达分析[J]. 中国农业科技导报, 2025, 27(7): 30-43. |
| [5] | 呼斯乐, 包玉龙, 图布新巴雅尔null, 陶际峰, 郭恩亮. 基于无人机高光谱和集成学习的春小麦叶绿素含量反演[J]. 中国农业科技导报, 2025, 27(6): 93-103. |
| [6] | 史硕, 冯宇, 李亮, 孟瑞, 章延泽, 杨秀荣. 印度梨形孢介导小麦抗纹枯病的转录组分析及关键基因筛选[J]. 中国农业科技导报, 2025, 27(5): 133-145. |
| [7] | 马蓓, 公杰, 杜银柯, 甘雨薇, 程蓉, 朱波, 易丽霞, 马锦绣, 高世庆. 小麦花粉孔发育相关TaINP1基因鉴定及表达分析[J]. 中国农业科技导报, 2025, 27(4): 22-35. |
| [8] | 陈宜新, 杨秀波, 田士军, 王聪, 白志英, 李存东, 张科. 陆地棉GhCOMT28对干旱胁迫的响应[J]. 中国农业科技导报, 2025, 27(4): 45-56. |
| [9] | 薛振宇, 张康康, 张元元, 闫强强, 姚立蓉, 张宏, 孟亚雄, 司二静, 李葆春, 马小乐, 王化俊, 汪军成. 优质抗旱小麦种质的筛选及功能基因检测[J]. 中国农业科技导报, 2025, 27(1): 35-49. |
| [10] | 孙宪印, 牟秋焕, 米勇, 吕广德, 亓晓蕾, 孙盈盈, 尹逊栋, 王瑞霞, 吴科, 钱兆国, 赵岩, 高明刚. 基于GT双标图对小麦新品系的分类评价[J]. 中国农业科技导报, 2024, 26(7): 14-24. |
| [11] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [12] | 赵刚, 王淑英, 李尚中, 张建军, 党翼, 王磊, 李兴茂, 程万莉, 周刚, 倪胜利, 樊廷录. 黄土旱塬区近40年降水对冬小麦耗水和产量的影响[J]. 中国农业科技导报, 2024, 26(3): 164-173. |
| [13] | 张宏, 李卫国, 张晓东, 卢必慧, 张琤琤, 李伟, 马廷淮. 基于HJ-1星和GF-1号影像融合特征提取冬小麦种植面积[J]. 中国农业科技导报, 2024, 26(2): 109-119. |
| [14] | 张景云, 关峰, 石博, 万新建. 小麦根系分泌物对苦瓜幼苗生长及土壤生物学环境的影响[J]. 中国农业科技导报, 2024, 26(2): 181-190. |
| [15] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||