




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (9): 79-91.DOI: 10.13304/j.nykjdb.2024.0208
谭华强1( ), 李丽平1, 铁曼曼2, 杨家勤1, 郑晓云1, 潘绍坤1, 唐有万1(
), 李丽平1, 铁曼曼2, 杨家勤1, 郑晓云1, 潘绍坤1, 唐有万1( )
)
收稿日期:2024-03-18
接受日期:2024-04-22
出版日期:2025-09-15
发布日期:2025-09-24
通讯作者:
唐有万
作者简介:谭华强 E-mail:307927595@qq.com;
基金资助:
Huaqiang TAN1( ), Liping LI1, Manman TIE2, Jiaqin YANG1, Xiaoyun ZHENG1, Shaokun PAN1, Youwan TANG1(
), Liping LI1, Manman TIE2, Jiaqin YANG1, Xiaoyun ZHENG1, Shaokun PAN1, Youwan TANG1( )
)
Received:2024-03-18
Accepted:2024-04-22
Online:2025-09-15
Published:2025-09-24
Contact:
Youwan TANG
摘要:
为深入研究紫色辣椒花青素降解的分子机制,选用3份紫色辣椒材料(HN191、HN192和HN005),对其花后30 d紫色未熟果实和60 d红色成熟果实进行转录组测序。结果表明,鉴定出7 104个共有的差异表达基因。GO和KEGG富集分析显示,这些共有差异表达基因主要富集在细胞壁、次生代谢、脂质代谢、碳水化合物代谢、氨基酸代谢相关的通路。进一步分析鉴定了3个MBW(MYB-bHLH-WD40)转录因子(包括1个MYB和2个WRKY)、3个过氧化物酶、2个β-葡萄糖苷酶编码基因;通过加权基因共表达网络分析(weighted gene co-expression network analysis,WGCNA)鉴定出1个MADS-box转录因子和1个NAC转录因子。实时荧光定量(quantitative real-time PCR,qRT-PCR)分析表明,这10个基因在红色成熟果实中的表达量均较紫色未熟果实上调,表明这10个候选基因可能与紫色辣椒果实的花青素降解有关。以上研究结果为紫色成熟辣椒的育种提供了潜在基因资源。
中图分类号:
谭华强, 李丽平, 铁曼曼, 杨家勤, 郑晓云, 潘绍坤, 唐有万. 紫色辣椒花青素降解相关基因的挖掘与分析[J]. 中国农业科技导报, 2025, 27(9): 79-91.
Huaqiang TAN, Liping LI, Manman TIE, Jiaqin YANG, Xiaoyun ZHENG, Shaokun PAN, Youwan TANG. Mining and Analysis of Genes Related to Anthocyanin Degradation in Purple Pepper[J]. Journal of Agricultural Science and Technology, 2025, 27(9): 79-91.
基因编号 Gene ID | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer(5’-3’) | 大小 Size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| T459_29804 | TGGAAGAGTTGAACCACCGA | GTGTCCTCGTCACCACTACA | 248 | 59 |
| T459_02736 | GGACCAACAACACACCAACA | CTTGGGTCACTGAAGCATCG | 249 | 59 |
| T459_27911 | AACGGGTTCTTGGGGCTAAT | GTGCATCGGTAGTAGCTCCT | 236 | 59 |
| T459_07162 | CTGCCAGTTGACTTGTTCGT | TGCAAAACGGTCACAGTGAG | 249 | 59 |
| T459_09393 | CTGCCAACACTCAAATCCCC | ATTTGCATCGCCACCAGAAG | 230 | 59 |
| T459_33197 | TTACCCAACAGTTGACCCGA | CAGCAGCCATTTTCTCCACA | 239 | 59 |
| T459_08317 | GTTGCAGCACAGAGAACCAA | ACCAAAGTCCCTGCTATGCT | 234 | 59 |
| T459_08320 | TGCTGGTTCTAGATTGCCCA | CTCCTTGCAGACCTGGTTCT | 233 | 59 |
| T459_27108 | TGCTTGGAGAGGATTTGGGA | TGTACACTTTGCTCCCCAGG | 244 | 59 |
| T459_12184 | ACCCGACTGATGAAGAGCTT | TCGCCTTCCAGTACCCATTT | 231 | 59 |
| T459_30033 | CCCTGTCCTGCTCACTGAAG | GTCACGTCCAGCAAGATCCA | 250 | 60 |
表1 候选基因的qRT-PCR分析引物
Table 1 Primers used in qRT-PCR for candidate genes
基因编号 Gene ID | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer(5’-3’) | 大小 Size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| T459_29804 | TGGAAGAGTTGAACCACCGA | GTGTCCTCGTCACCACTACA | 248 | 59 |
| T459_02736 | GGACCAACAACACACCAACA | CTTGGGTCACTGAAGCATCG | 249 | 59 |
| T459_27911 | AACGGGTTCTTGGGGCTAAT | GTGCATCGGTAGTAGCTCCT | 236 | 59 |
| T459_07162 | CTGCCAGTTGACTTGTTCGT | TGCAAAACGGTCACAGTGAG | 249 | 59 |
| T459_09393 | CTGCCAACACTCAAATCCCC | ATTTGCATCGCCACCAGAAG | 230 | 59 |
| T459_33197 | TTACCCAACAGTTGACCCGA | CAGCAGCCATTTTCTCCACA | 239 | 59 |
| T459_08317 | GTTGCAGCACAGAGAACCAA | ACCAAAGTCCCTGCTATGCT | 234 | 59 |
| T459_08320 | TGCTGGTTCTAGATTGCCCA | CTCCTTGCAGACCTGGTTCT | 233 | 59 |
| T459_27108 | TGCTTGGAGAGGATTTGGGA | TGTACACTTTGCTCCCCAGG | 244 | 59 |
| T459_12184 | ACCCGACTGATGAAGAGCTT | TCGCCTTCCAGTACCCATTT | 231 | 59 |
| T459_30033 | CCCTGTCCTGCTCACTGAAG | GTCACGTCCAGCAAGATCCA | 250 | 60 |
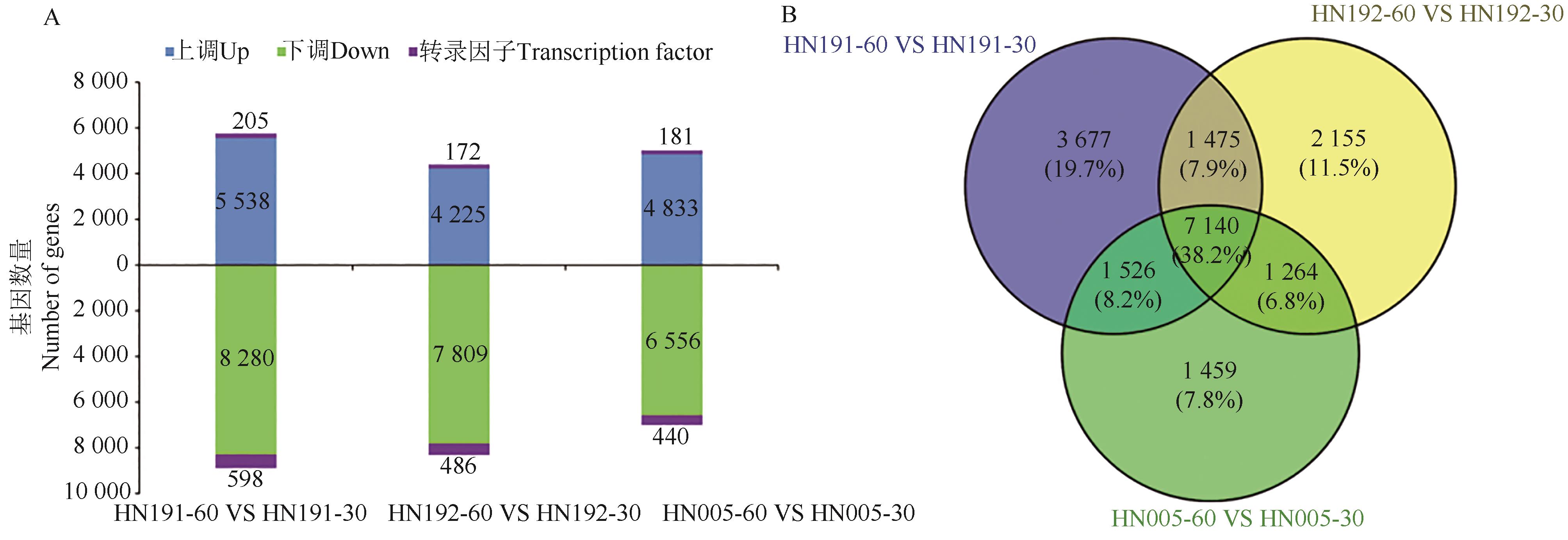
图1 不同品种30与60 d果实发育间的差异表达基因A:差异表达基因直方图; B:差异表达基因的维恩图
Fig. 1 DEGs between fruits of 30 and 60 d from different varietiesA:Histogram of differentially expressed genes; B. Venn plot of differentially expressed genes
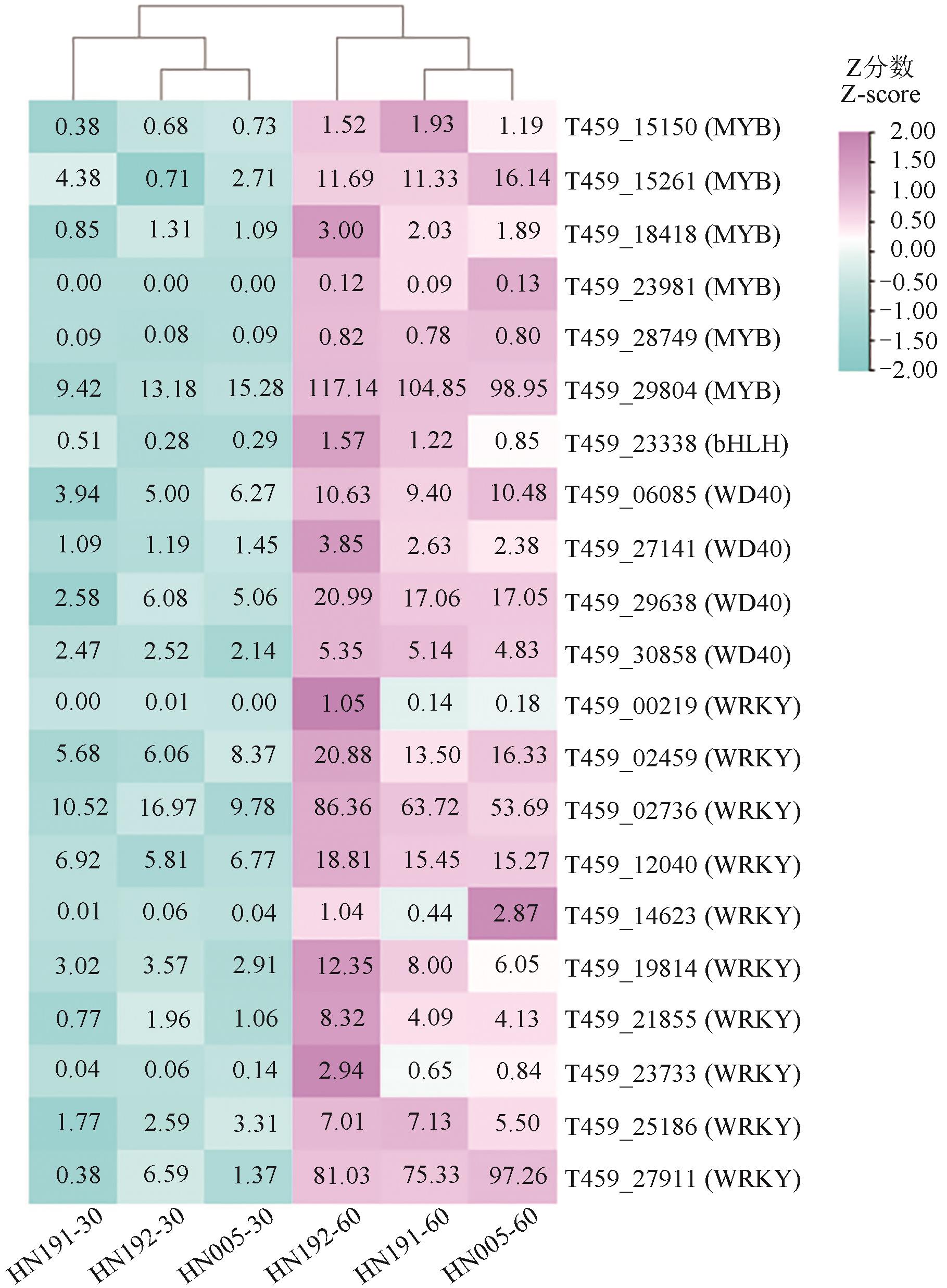
图3 共有差异表达基因中的MBW转录因子热图注:框中的数值代表FPKM值。
Fig. 3 Heatmap of MBW transcription factors predicted from common DEGsNote:Number in the box indicates FPKM value.
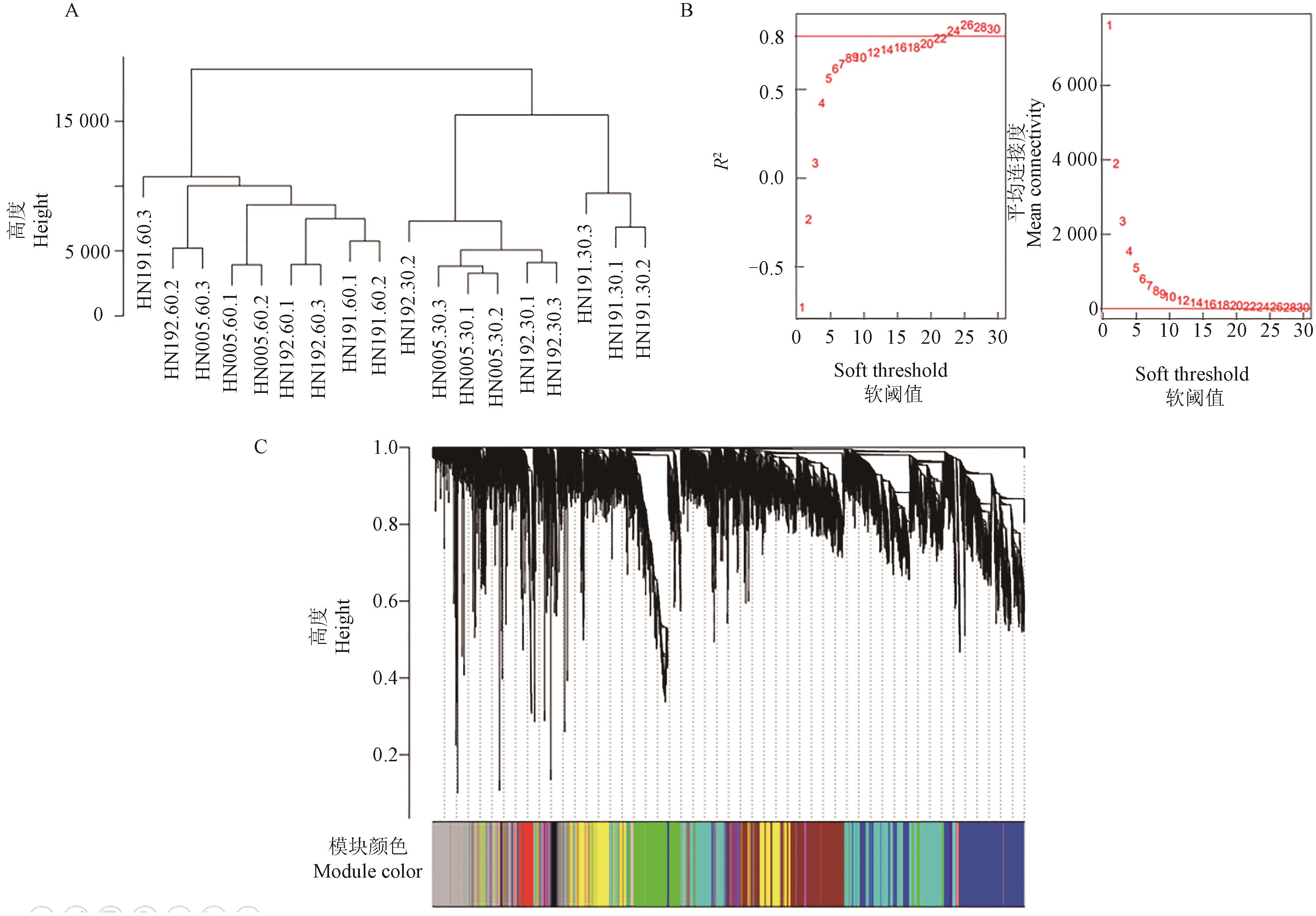
图6 加权共表达网络构建A:18个样本的聚类图;B:软阈值筛选;C:基因聚类和模块切割
Fig. 6 Construction of a weighted co-expression networkA:Cluster dendrogram of 18 samples;B:Soft threshold selection;C:Gene clustering and module cutting
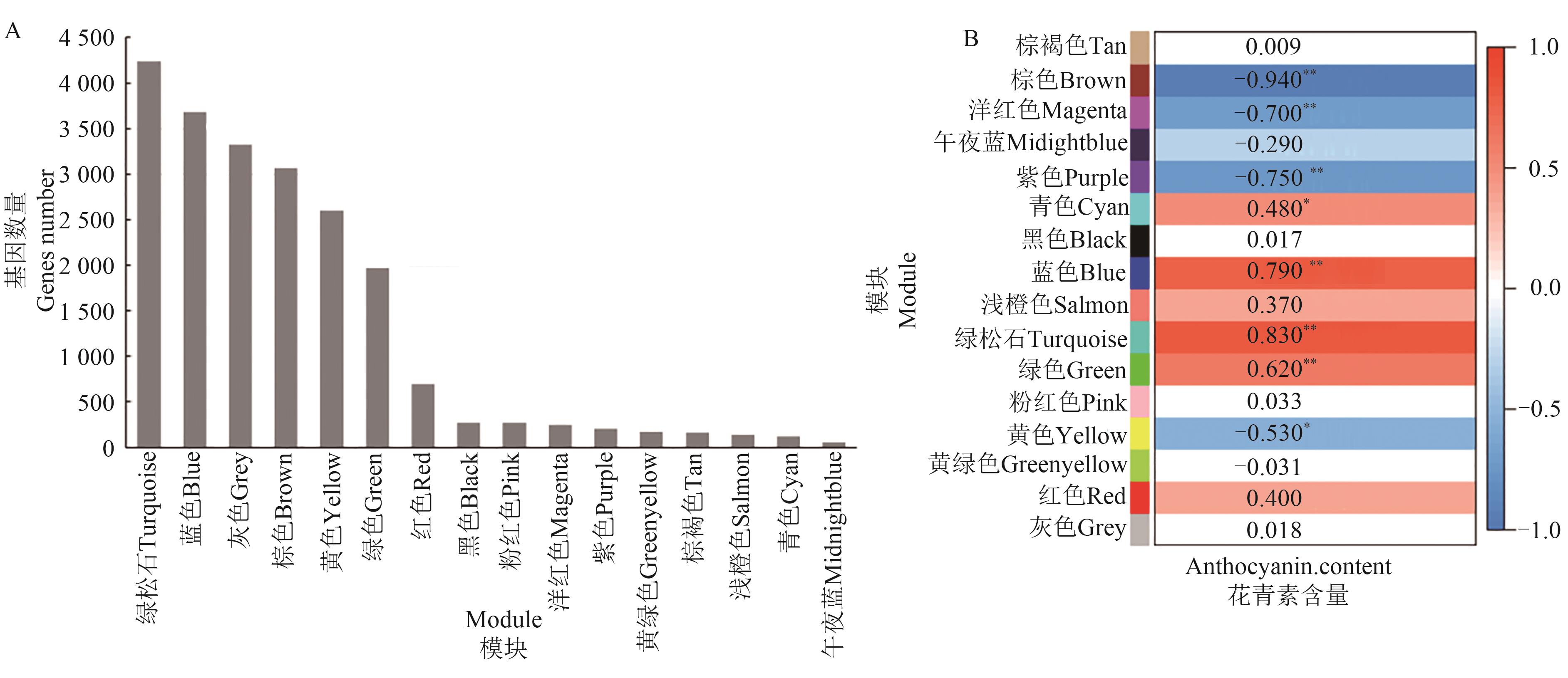
图7 WGCNA模块分析A:16个模块所包含的基因数量统计;B:16个模块与花青素含量的相关性热图。*和**分别表示在P<0.05和P<0.01水平显著相关
Fig. 7 Analysis of WGCNA modulesA: Statistics of gene numbers in 16 modules; B: Relationship heatmap of 16 modules with anthocyanin content. * and ** indicate significant correlations at P<0.05 and P<0.01 levels, respectively
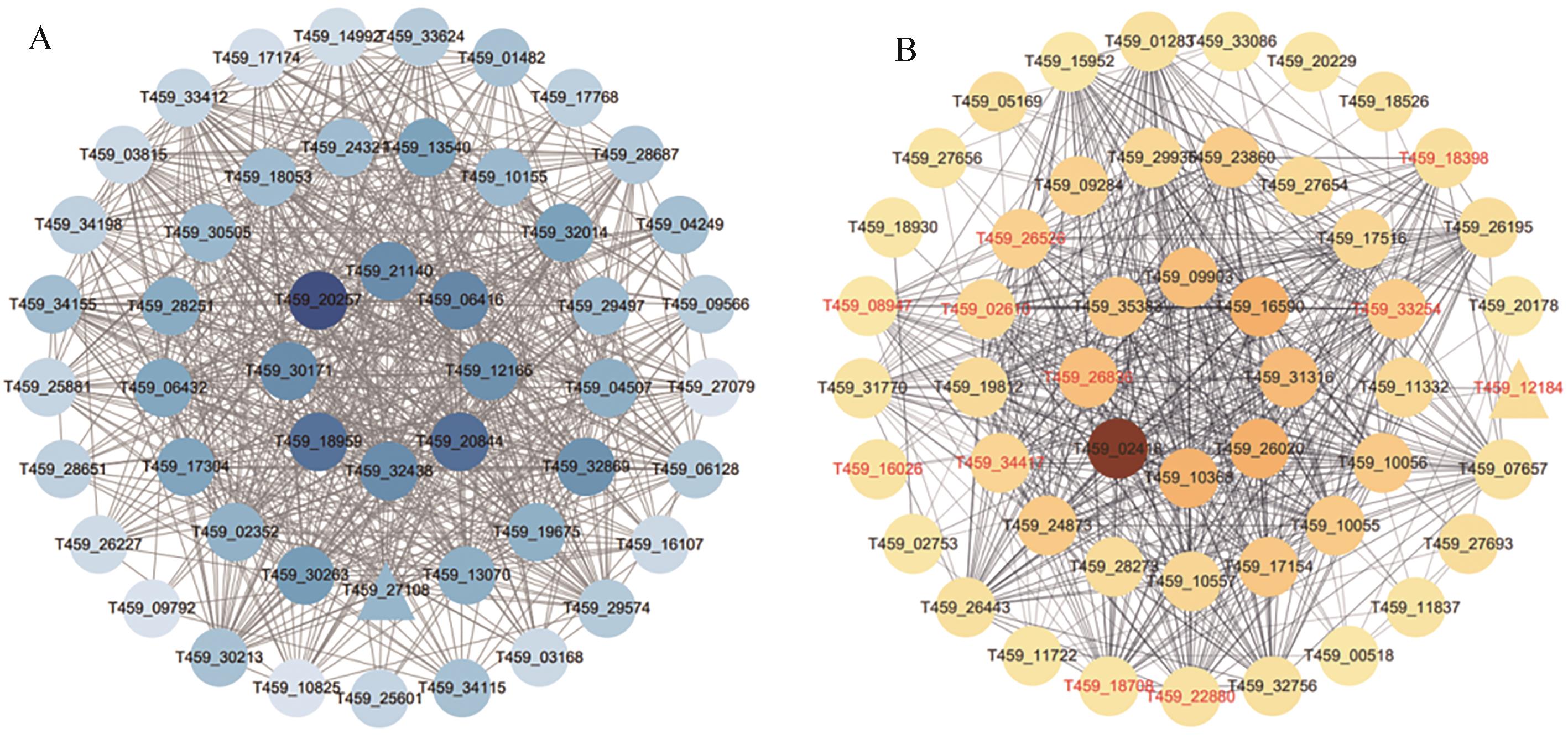
图8 WGCNA网络Hub基因分析A: WGCNA得到的蓝色模块degree前50个基因网络图。颜色深浅代表Degree值的大小,颜色越深代表Degree值越高,三角形节点代表转录因子;B:共有差异表达基因PPI网络Degree前50个基因网络图。红色基因名代表与蓝色模块共有的基因,颜色深浅代表Degree值的大小,颜色越深代表degree值越高。三角形节点代表转录因子
Fig. 8 Analysis of hub genes in WGCNA networkA: Network diagram of degree top 50 genes in the blue module obtained by WGCNA. The color shade on the diagram represents the Degree value, with darker colors indicating higher values and triangular node in the diagram represents transcription factor;B: Network diagram of Degree top 50 genes in PPI network of common DEGs. The red gene names indicate genes share with the blue module, the color shade represents the Degree value, with darker colors indicating higher values. Triangular node in this diagram represents transcription factor
| [1] | PERRY L, DICKAU R, ZARRILLO S,et al..Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp.L.) in the Americas [J]. Science, 2007, 315(5814):986-988. |
| [2] | 邹学校,马艳青,戴雄泽,等.辣椒在中国的传播与产业发展[J].园艺学报,2020,47(9):1715-1726. |
| ZOU X X, MA Y Q, DAI X Z, et al.. Spread and industry development of pepper in China [J]. Acta Hortic. Sin., 2020, 47(9):1715-1726. | |
| [3] | STOMMEL J R, LIGHTBOURN G J, WINKEL B S, et al.. Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum [J]. J. Am. Soc. Hortic. Sci., 2009, 134(2): 244-251. |
| [4] | CHALKER-SCOTT L. Environmental significance of anthocyanins in plant stress responses [J]. Photochem. Photobiol., 1999, 70(1):1-9. |
| [5] | DE PASCUAL-TERESA S, MORENO D A, GARCÍA-VIGUERA C.Flavanols and anthocyanins in cardiovascular health:a review of current evidence [J]. Int. J. Mol. Sci., 2010,11(4):1679-1703. |
| [6] | LIU Y, TIKUNOV Y, SCHOUTEN R E,et al.. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables:a review [J/OL].Front.Chem.,2018,6:52 [2024-02-20]. . |
| [7] | JAAKOLA L.New insights into the regulation of anthocyanin biosynthesis in fruits [J]. Trends Plant Sci., 2013, 18(9):477-483. |
| [8] | BOROVSKY Y, OREN-SHAMIR M, OVADIA R, et al.. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia [J]. Theor. Appl. Genet., 2004, 109(1):23-29. |
| [9] | JUNG S, VENKATESH J, KANG M Y,et al..A non-LTR retrotransposon activates anthocyanin biosynthesis by regulating a MYB transcription factor in Capsicum annuum [J/OL].Plant Sci.,2019,287:110181 [2024-02-20]. . |
| [10] | LIU J Q, AI X Y, WANG Y H, et al..Fine mapping of the Ca3GT gene controlling anthocyanin biosynthesis in mature unripe fruit of Capsicum annuum L. [J]. Theor. Appl. Genet., 2020,133(9):2729-2742. |
| [11] | WANG G, CHEN B, DU H,et al..Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA [J/OL].PLoS One,2018,13(9):e0204690 [2024-02-20]. . |
| [12] | TANG B Y, LI L, HU Z L, et al.. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper [J]. J. Agric. Food Chem., 2020,68(43):12152-12163. |
| [13] | LIU Y H, LYU J H, LIU Z B,et al..Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.) [J/OL]. Food Chem., 2020, 306:125629 [2024-02-20]. . |
| [14] | MENG Y, ZHANG H, FAN Y, et al.. Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum) [J/OL]. BMC Plant Biol., 2022,22(1):358 [2024-02-20]. . |
| [15] | WANG Y, LIU S, WANG H, et al.. Identification of the regulatory genes of UV-B-induced anthocyanin biosynthesis in pepper fruit [J/OL]. Int. J. Mol. Sci., 2022,23(4):1960 [2024-02-20]. . |
| [16] | ZHOU Y, MUMTAZ MALI, ZHANG Y H, et al.. Response of anthocyanin biosynthesis to light by strand-specific transcriptome and miRNA analysis in Capsicum annuum [J/OL]. BMC Plant Biol., 2022,22(1):79 [2024-02-20]. . |
| [17] | TAN H, LI L, TIE M, et al.. Transcriptome analysis of green and purple fruited pepper provides insight into novel regulatory genes in anthocyanin biosynthesis [J/OL]. PeerJ, 2024,12:e16792 [2024-02-20]. . |
| [18] | PASSERI V, KOES R, QUATTROCCHIO F M. New challenges for the design of high value plant products:stabilization of anthocyanins in plant vacuoles [J/OL]. Front. Plant Sci., 2016,7:153 [2024-02-20]. . |
| [19] | KOES R, VERWEIJ W, QUATTROCCHIO F.Flavonoids:a colorful model for the regulation and evolution of biochemical pathways [J]. Trends Plant Sci., 2005,10(5):236-242. |
| [20] | VERWEIJ W, SPELT C, DI SANSEBASTIANO G P, et al.. An H+P-ATPase on the tonoplast determines vacuolar pH and flower colour [J]. Nat. Cell Biol., 2008,10:1456-1462. |
| [21] | FARACO M, SPELT C, BLIEK M, et al..Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color [J]. Cell Rep., 2014,6(1):32-43. |
| [22] | DE VLAMING P, SCHRAM A W, WIERING H. Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrid [J]. Theor. Appl. Genet., 1983, 66(3):271-278. |
| [23] | DE VLAMING P, VAN EEKERES J E M, WIERING H. A gene for flower colour fading in Petunia hybrid [J]. Theor. Appl.Genet., 1982, 61(1):41-46. |
| [24] | CHASSY A W, BUESCHL C, LEE H, et al.. Tracing flavonoid degradation in grapes by MS filtering with stable isotopes [J]. Food Chem., 2015,166:448-455. |
| [25] | OREN-SHAMIR M.Does anthocyanin degradation play a significant role in determining pigment concentration in plants? [J]. Plant Sci., 2009, 177(4):310-316. |
| [26] | FANG F, ZHANG X L, LUO H H, et al.. An intracellular laccase is responsible for epicatechin-mediated anthocyanin degradation in Litchi fruit pericarp [J]. Plant Physiol., 2015, 169(4):2391-2408. |
| [27] | MOVAHED N, PASTORE C, CELLINI A, et al.. The grapevine VviPrx31 peroxidase as a candidate gene involved in anthocyanin degradation in ripening berries under high temperature [J]. J. Plant Res., 2016, 129(3):513-526. |
| [28] | ZIPOR G, DUARTE P, CARQUEIJEIRO I,et al..In planta anthocyanin degradation by a vacuolar class Ⅲ peroxidase in Brunfelsia calycina flowers [J]. N. Phytol., 2015, 205(2):653-665. |
| [29] | WANG L S, BURHENNE K, KRISTENSEN B K, et al.. Purification and cloning of a Chinese red radish peroxidase that metabolise pelargonidin and forms a gene family in Brassicaceae [J]. Gene, 2004, 343(2):323-335. |
| [30] | YAMADA Y, NAKAYAMA M, SHIBATA H, et al.. Anthocyanin production and enzymatic degradation during the development of dark purple and lilac paprika fruit [J]. J. Am. Soc. Hortic. Sci., 2019, 144(5): 329-338. |
| [31] | SAKAMURA S, OBATA Y.Anthocyanase and anthocyanins occurring in eggplant,Solanum melongena L.(I) [J]. Agric. Biol. Chem., 1961, 25(10):750-756. |
| [32] | BARBAGALLO R N, PALMERI R, FABIANO S, et al.. Characteristic of β-glucosidase from Sicilian blood oranges in relation to anthocyanin degradation [J]. Enzyme Microb. Technol., 2007, 41(5):570-575. |
| [33] | DONG B, LUO H, LIU B, et al.. BcXyl,a β-xylosidase isolated from Brunfelsia calycina flowers with anthocyanin-β-glycosidase activity [J/OL]. Investig. N. Drugs, 2019,20(6):E1423 [2024-02-20]. . |
| [34] | ZHAO G, XIANG F, ZHANG S, et al.. PbLAC4-like,activated by PbMYB26,related to the degradation of anthocyanin during color fading in pear [J/OL]. BMC Plant Biol., 2021,21(1):469 [2024-02-20]. . |
| [35] | AZA-GONZÁLEZ C, HERRERA-ISIDRÓN L, NÚÑEZ-PALENIUS H G,et al..Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development [J]. Biol. Plant., 2013, 57(1):49-55. |
| [36] | CHEN S F, ZHOU Y Q, CHEN Y R, et al.. Fastp:an ultra-fast all-in-one FASTQ preprocessor [J]. Bioinformatics, 2018,34(17):884-890. |
| [37] | KIM D, LANGMEAD B, SALZBERG S L. HISAT:a fast spliced aligner with low memory requirements [J]. Nat. Methods, 2015,12:357-360. |
| [38] | KIM S, PARK M, YEOM S I, et al.. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species [J]. Nat. Genet., 2014, 46:270-278. |
| [39] | PERTEA M, KIM D, PERTEA G M,et al..Transcript-level expression analysis of RNA-seq experiments with HISAT,StringTie and Ballgown [J]. Nat. Protoc., 2016,11:1650-1667. |
| [40] | VARET H, BRILLET-GUÉGUEN L, COPPÉE J Y,et al..SARTools:a DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data [J/OL].PLoS One, 2016,11(6):e0157022 [2024-02-20]. . |
| [41] | CHEN C J, WU Y, LI J W, et al.. TBtools-II:a “one for all, all for one” bioinformatics platform for biological big-data mining [J].Mol. Plant, 2023,16(11):1733-1742. |
| [42] | ZHENG Y, JIAO C, SUN H, et al.. ITAK:a program for Genome-wide prediction and classification of plant transcription factors,transcriptional regulators,and protein kinases [J]. Mol. Plant, 2016,9(12):1667-1670. |
| [43] | LIVAK K J, SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001,25(4):402-408. |
| [44] | TOGNOLLI M, PENEL C, GREPPIN H,et al..Analysis and expression of the class Ⅲ peroxidase large gene family in Arabidopsis thaliana [J]. Gene, 2002,288(1/2):129-138. |
| [45] | LANGFELDER P, HORVATH S. WGCNA:an R package for weighted correlation network analysis [J/OL].BMC Bioinf.,2008,9:559 [2024-02-20]. . |
| [46] | SEYMOUR G B, ØSTERGAARD L, CHAPMAN N H,et al..Fruit development and ripening [J]. Annu. Rev. Plant Biol., 2013,64:219-241. |
| [47] | GROSS K C, WATADA A E, KANG M S, et al.. Biochemical changes associated with the ripening of hot pepper fruit [J]. Physiol. Plant., 1986, 66(1):31-36. |
| [48] | PRIYA SETHU K M, PRABHA T N, THARANATHAN R N. Post-harvest biochemical changes associated with the softening phenomenon in Capsicum annuum fruits [J].Phytochemistry,1996,42(4):961-966. |
| [49] | JANG Y K, JUNG E S, LEE H A,et al..Metabolomic characterization of hot pepper (Capsicum annuum “CM334”) during fruit development [J]. J. Agric. Food Chem., 2015,63(43):9452-9460. |
| [50] | LIU Z, LYU J, ZHANG Z, et al.. Integrative transcriptome and proteome analysis identifies major metabolic pathways involved in pepper fruit development [J].J. Proteome Res., 2019,18(3):982-994. |
| [51] | SONG J, SUN B, CHEN C, et al.. An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis [J]. Plant J., 2023,115(3):724-741. |
| [52] | TAO H, GAO F, LINYING LI,et al..WRKY33 negatively regulates anthocyanin biosynthesis and cooperates with PHR1 to mediate acclimation to phosphate starvation [J/OL]. Plant Commun., 2024,5(5):100821 [2024-02-20]. . |
| [53] | DEVAIAH B N, KARTHIKEYAN A S, RAGHOTHAMA K G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis [J].Plant Physiol., 2007,143(4):1789-1801. |
| [54] | DUAN S W, WANG J J, GAO C H, et al..Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana [J]. Plant Sci., 2018,268:47-53. |
| [55] | MAO Z L, JIANG H Y, WANG S,et al..The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple [J/OL]. Plant Sci., 2021,306:110848 [2024-02-20]. . |
| [56] | ZHANG Z Q, PANG X Q, JI Z L, et al.. Role of anthocyanin degradation in litchi pericarp browning [J]. Food Chem., 2001,75(2):217-221. |
| [57] | NORMAN C, RUNSWICK M, POLLOCK R, et al.. Isolation and properties of cDNA clones encoding SRF,a transcription factor that binds to the c-fos serum response element [J]. Cell, 1988,55(6):989-1003. |
| [58] | SAEDLER H, BECKER A, WINTER K U,et al.. MADS-box genes are involved in floral development and evolution [J]. Acta Biochim. Pol., 2001, 48(2):351-358. |
| [59] | LI C, WANG Y, XU L, et al.. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis [J/OL]. Front. Plant Sci., 2016,7:1390 [2024-02-20]. . |
| [60] | LU W J, CHEN J X, REN X C, et al.. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening [J]. Sci. Hortic., 2018, 227:124-131. |
| [61] | MA G Y, ZOU Q C, SHI X H, et al.. Ectopic expression of the AaFUL1 gene identified in Anthurium andraeanum affected floral organ development and seed fertility in tobacco [J]. Gene, 2019, 696:197-205. |
| [62] | WU R, WANG T, MCGIE T,et al..Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals,but has no effect on vegetative growth,dormancy,or flowering time [J]. J. Exp.Bot., 2014, 65(17):4985-4995. |
| [63] | QI F T, LIU Y T, LUO Y L, et al.. Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets [J/OL]. Hortic. Res., 2022,9:uhac071 [2024-02-20]. . |
| [64] | OOKA H, SATOH K, DOI K, et al.. Comprehensive analysis of nac family genes in Oryza sativa and Arabidopsis thaliana [J].DNA Res., 2003, 10(6):239-247. |
| [65] | OLSEN A N, ERNST H A, LEGGIO L L, et al.. NAC transcription factors:structurally distinct,functionally diverse [J]. Trends Plant Sci., 2005, 10(2):79-87. |
| [66] | NURUZZAMAN M, SHARONI A M, KIKUCHI S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants [J/OL]. Front. Microbiol., 2013,4:248 [2024-02-20]. . |
| [67] | MAHMOOD K, XU Z, EL-KEREAMY A, et al.. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses [J/OL]. Front. Plant Sci., 2016,7:1548 [2024-02-20]. . |
| [68] | WU A H, ALLU A D, GARAPATI P, et al.. JUNGBRUNNEN1,a reactive oxygen species-responsive NAC transcription factor,regulates longevity in Arabidopsis [J]. Plant Cell, 2012,24(2):482-506. |
| [69] | WANG J F, LIAN W R, CAO Y Y, et al.. Overexpression of BoNAC019,a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis [J/OL]. Sci. Rep., 2018, 8:13349 [2024-02-20]. . |
| [1] | 刘雪晴, 王静, 阳宜, 吴慧琴, 王延宏, 王麓尧, 路佳伟, 张凯璇, 翟源, 成妍. 外源乙烯利对色素辣椒脱叶及产量的影响[J]. 中国农业科技导报, 2025, 27(8): 36-46. |
| [2] | 雷刚, 方荣, 周坤华, 陈学军, 袁欣捷, 黄月琴, 李歌歌, 谢媛媛, 宋小民. 辣椒种质资源苗期耐热性综合评价体系的建立[J]. 中国农业科技导报, 2025, 27(3): 60-70. |
| [3] | 张余莽, 陈贵娟, 常洪艳, 王永恒, 刘淑霞, 应允秀. 水分胁迫下新型土壤保水剂对玉米苗期发育的影响[J]. 中国农业科技导报, 2025, 27(1): 201-210. |
| [4] | 刘徐冬雨, 郭潇潇, 付晨青, 韩蕊, 李国辉, 王秀萍. 基于RGB和CIELab预测紫苏叶片花青素含量[J]. 中国农业科技导报, 2024, 26(7): 103-110. |
| [5] | 谢勇俊, 潘小卓, 陈福慧, 尹凯波, 金嘉悦, 王一兵. 人参酚酸类自毒物质降解菌的筛选鉴定及生防研究[J]. 中国农业科技导报, 2024, 26(7): 147-155. |
| [6] | 姚建民, 马俊奎, 王忠祥, 毕昕媛, 李瑞珍, 杨瑞平, 刘钊, 郭丰辉. 全生物降解渗水地膜在大豆-玉米带状复合种植中的应用效果研究[J]. 中国农业科技导报, 2023, 25(9): 178-185. |
| [7] | 张德俊, 张小明, 吴荻, 林蜀云, 张太华, 徐卫平. 基于DEM-CFD耦合的辣椒清选仿真研究[J]. 中国农业科技导报, 2023, 25(7): 87-96. |
| [8] | 李瑞珍, 姚建民, 王忠祥, 高凤翔, 窦贵新, 杨瑞平, 刘钊, 张继, 张振宇. 全生物降解渗水地膜覆盖冬播谷子产量结构关系分析[J]. 中国农业科技导报, 2023, 25(5): 185-191. |
| [9] | 柳春雨, 门丽娜, 连雅琴, 张宇宏, 张志伟, 张伟. 大蜡螟幼虫肠道微生物聚乙烯降解酶基因的表达及性质分析[J]. 中国农业科技导报, 2023, 25(3): 132-139. |
| [10] | 杨文竹, 陈茹梅. 花青素玉米的育种进展[J]. 中国农业科技导报, 2022, 24(8): 18-24. |
| [11] | 吴玉洪, 郭荣君, 马桂珍, 李世东. 嗜吡啶红球菌Rp3生长及降解粪臭素特性研究[J]. 中国农业科技导报, 2022, 24(6): 82-89. |
| [12] | 刘璐, 陶秀萍, 宋建超, 尚斌, 徐文倩, 董红敏, 蔡阳扬. 微生物燃料电池处理奶牛场污水运行效果与产电性能试验研究[J]. 中国农业科技导报, 2022, 24(4): 134-143. |
| [13] | 王玉, 李春光, 刘欢, 张月华, 冯晓民, 李耀光, 李怀奇, 景延秋, 孙觅. 烤烟叶片叶绿体超微结构与质体色素降解产物的关系初探[J]. 中国农业科技导报, 2022, 24(3): 67-76. |
| [14] | 范文华, 戴建军, 张树山, 孙玲伟, 吴彩凤, 张德福. 大豆卵磷脂稀释液中添加原花青素对山羊冻精效果的影响#br#[J]. 中国农业科技导报, 2021, 23(9): 78-86. |
| [15] | 程云霞§, 吴慧§, 刘迁杰, 时振宇, 贾凯, 陈奕琳, 俞安伟, 申金秀. 不同剂量保水剂对袋式复合沙培辣椒生长发育及品质的影响[J]. 中国农业科技导报, 2021, 23(8): 55-62. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||