




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (2): 27-37.DOI: 10.13304/j.nykjdb.2022.0612
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Ting LIANG1( ), Jinghong ZUO2, Qing LU1, Dong YANG2, Yimiao TANG2, Chunman GUO2(
), Jinghong ZUO2, Qing LU1, Dong YANG2, Yimiao TANG2, Chunman GUO2( ), Dezhou WANG2(
), Dezhou WANG2( ), Weiwei WANG2(
), Weiwei WANG2( )
)
Received:2022-07-19
Accepted:2022-09-28
Online:2023-02-15
Published:2023-05-17
Contact:
Chunman GUO,Dezhou WANG,Weiwei WANG
梁婷1( ), 左静红2, 陆青1, 杨东2, 唐益苗2, 郭春曼2(
), 左静红2, 陆青1, 杨东2, 唐益苗2, 郭春曼2( ), 汪德州2(
), 汪德州2( ), 王伟伟2(
), 王伟伟2( )
)
通讯作者:
郭春曼,汪德州,王伟伟
作者简介:梁婷 E-mail:1577033136@qq.com
基金资助:CLC Number:
Ting LIANG, Jinghong ZUO, Qing LU, Dong YANG, Yimiao TANG, Chunman GUO, Dezhou WANG, Weiwei WANG. Identification and Expression Analysis Under Abiotic Stress of IQM Gene Family in Wheat (Triticum aestivum L.)[J]. Journal of Agricultural Science and Technology, 2023, 25(2): 27-37.
梁婷, 左静红, 陆青, 杨东, 唐益苗, 郭春曼, 汪德州, 王伟伟. 小麦IQM基因家族鉴定及非生物胁迫下表达分析[J]. 中国农业科技导报, 2023, 25(2): 27-37.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2022.0612
| 基因 Gene | 正向引物 Forward primer (5’–3’) | 反向引物 Reverse primer (5’–3’) |
|---|---|---|
| TaIQM1 | GACGATTGGTGGTCAAAGATGG | ACGCCGTCTATCTCAACTTCTGTC |
| TaIQM2 | TCGTTTCAGCACTCCAGTTTCC | TTGACATCGGTGAGGCTAACGT |
| TaIQM3 | CAAGGGCAGTGCGTCAAGTA | TGGAGTCATCCGAAGTGTGC |
| TaIQM4 | GGAGATGTTCACAAGGCTGGATTC | GGGTAGTCCCTGACGCAAATGAT |
| TaIQM5 | ATGGAGTTCTGAAGGCTATCTGGC | TTCTTCGGGTTTGTCGGCATC |
| TaIQM6 | GTGCCAAGGGAGAAGGTCATT | GGTAGTCCCTGACGCAAATGAT |
| TaIQM7 | CGAGCCTACTGAAACGGAAGAAC | CCATTTGAAGGACGGGAGACTG |
| TaIQM8 | TCAAGAACTGGGAGGCGGAG | AGACGAAGTGCGGCTTGGAC |
| TaIQM9 | CGAGAGGGCAAGGTATGAGGTTA | ACCCTTCTTCTTCTGCCCGAT |
| TaIQM10 | AACTCGGACAGGCCATTTGT | TAGAGCTCACCGCATCGAAC |
| TaIQM11 | CCATAAGCACAACCACAGCCT | CAACAATGGACGACAAGCACAG |
| TaIQM12 | CTCGCCAGCAGATGATGACA | AAGGACCCTTTTCTCGCAGG |
| TaIQM13 | GCTCATCCACCAAGGAAGACTAC | GGTACCATGTTGGAAGGCAGT |
| TaIQM14 | GGAACCCTGAAGGCTATTTGG | TGGTTCCTCCCGTTTCTCTGT |
| TaIQM15 | TAATGTCCGTGTGGAGCAAGCA | CGCTTGCATTCTGAGCTCTATCG |
| TaIQM16 | GTGGTCACTACCGCCCTACA | CCTCTGCTGGGCTCATCTTC |
| TaIQM17 | TGGAGCAAGCGAGACCAACCTA | GGCCGAGCTGATAGGATTTTGAC |
| TaIQM18 | GAGAGGACTATGAGGTCGTGATTG | CCATTTTCAGCAACCAGCCT |
| TaIQM19 | CCCAACTGATTCTCCCTTCCAAC | CCACACGGAGATTATCGTTGACCT |
| TaIQM20 | CACAACTGATTCTCCCTTCCAAC | ATTCCCAGTGCTCCATTTCAG |
| TaIQM21 | AGTGGACATTACAAACCAAGTGCG | ATTGTGGAGGATTGGATGGCA |
| TaIQM22 | TGGACATTACAAACCGAGTGCG | AGCAGCAGTAGGGTTCTGTTTGGT |
| TaIQM23 | CGTCAGCCTCAACGCATCAA | GGTTATGCCCGTACCTATGTCTTG |
| 18S | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
Table 1 qRT-PCR primers
| 基因 Gene | 正向引物 Forward primer (5’–3’) | 反向引物 Reverse primer (5’–3’) |
|---|---|---|
| TaIQM1 | GACGATTGGTGGTCAAAGATGG | ACGCCGTCTATCTCAACTTCTGTC |
| TaIQM2 | TCGTTTCAGCACTCCAGTTTCC | TTGACATCGGTGAGGCTAACGT |
| TaIQM3 | CAAGGGCAGTGCGTCAAGTA | TGGAGTCATCCGAAGTGTGC |
| TaIQM4 | GGAGATGTTCACAAGGCTGGATTC | GGGTAGTCCCTGACGCAAATGAT |
| TaIQM5 | ATGGAGTTCTGAAGGCTATCTGGC | TTCTTCGGGTTTGTCGGCATC |
| TaIQM6 | GTGCCAAGGGAGAAGGTCATT | GGTAGTCCCTGACGCAAATGAT |
| TaIQM7 | CGAGCCTACTGAAACGGAAGAAC | CCATTTGAAGGACGGGAGACTG |
| TaIQM8 | TCAAGAACTGGGAGGCGGAG | AGACGAAGTGCGGCTTGGAC |
| TaIQM9 | CGAGAGGGCAAGGTATGAGGTTA | ACCCTTCTTCTTCTGCCCGAT |
| TaIQM10 | AACTCGGACAGGCCATTTGT | TAGAGCTCACCGCATCGAAC |
| TaIQM11 | CCATAAGCACAACCACAGCCT | CAACAATGGACGACAAGCACAG |
| TaIQM12 | CTCGCCAGCAGATGATGACA | AAGGACCCTTTTCTCGCAGG |
| TaIQM13 | GCTCATCCACCAAGGAAGACTAC | GGTACCATGTTGGAAGGCAGT |
| TaIQM14 | GGAACCCTGAAGGCTATTTGG | TGGTTCCTCCCGTTTCTCTGT |
| TaIQM15 | TAATGTCCGTGTGGAGCAAGCA | CGCTTGCATTCTGAGCTCTATCG |
| TaIQM16 | GTGGTCACTACCGCCCTACA | CCTCTGCTGGGCTCATCTTC |
| TaIQM17 | TGGAGCAAGCGAGACCAACCTA | GGCCGAGCTGATAGGATTTTGAC |
| TaIQM18 | GAGAGGACTATGAGGTCGTGATTG | CCATTTTCAGCAACCAGCCT |
| TaIQM19 | CCCAACTGATTCTCCCTTCCAAC | CCACACGGAGATTATCGTTGACCT |
| TaIQM20 | CACAACTGATTCTCCCTTCCAAC | ATTCCCAGTGCTCCATTTCAG |
| TaIQM21 | AGTGGACATTACAAACCAAGTGCG | ATTGTGGAGGATTGGATGGCA |
| TaIQM22 | TGGACATTACAAACCGAGTGCG | AGCAGCAGTAGGGTTCTGTTTGGT |
| TaIQM23 | CGTCAGCCTCAACGCATCAA | GGTTATGCCCGTACCTATGTCTTG |
| 18S | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
基因 Gene | 基因号 Gene ID | 染色体定位 Chromosome localization | 氨基酸数 Number of amino acids | 相对分子量 Molecular weight/Da | 等电点 Point isoelectric | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TaIQM1 | TraesCS1A02G125400.1 | Chr1A: 149352287-149356056 | 517 | 57 811.59 | 7.68 | 细胞核Nucleus |
| TaIQM2 | TraesCS1B02G143800.1 | Chr1B: 195555337-195560543 | 580 | 65 214.18 | 7.69 | 细胞核 Nucleus |
| TaIQM3 | TraesCS1D02G128400.1 | Chr1D: 141545585-141547817 | 424 | 47 667.67 | 9.49 | 细胞核Nucleus |
| TaIQM4 | TraesCS2A02G154900.1 | Chr2A: 102130189-102132879 | 564 | 63 624.94 | 6.47 | 细胞核Nucleus |
| TaIQM5 | TraesCS2B02G180000.1 | Chr2B: 154592307-154595019 | 572 | 64 351.52 | 6.36 | 细胞核Nucleus |
| TaIQM6 | TraesCS2D02G160200.1 | Chr2D: 103126893-103129599 | 571 | 64 331.63 | 6.26 | 细胞核Nucleus |
| TaIQM7 | TraesCS3A02G206400.1 | Chr3A: 363415357-363419189 | 556 | 62 188.59 | 9.13 | 细胞核Nucleus |
| TaIQM8 | TraesCS3B02G238500.1 | Chr3B: 373842628-373844908 | 541 | 60 758.12 | 9.07 | 细胞核Nucleus |
| TaIQM9 | TraesCS3D02G209200.1 | Chr3D: 276574646-276577885 | 530 | 59 496.51 | 8.97 | 细胞核Nucleus |
| TaIQM10 | TraesCS4A02G021000.1 | Chr4A: 14261744-14265762 | 610 | 67 879.37 | 9.60 | 细胞核Nucleus |
| TaIQM11 | TraesCS4B02G282600.2 | Chr4B: 565861371-565865127 | 669 | 74 779.20 | 9.41 | 细胞核Nucleus |
| TaIQM12 | TraesCS4D02G281600.1 | Chr4D: 452795390-452798654 | 613 | 68 128.53 | 9.67 | 细胞核Nucleus |
| TaIQM13 | TraesCS5A02G129600.1 | Chr5A: 290924878-290930090 | 461 | 51 276.55 | 6.88 | 细胞核Nucleus |
| TaIQM14 | TraesCS5A02G375700.1 | Chr5A: 573606336-573609441 | 534 | 59 693.35 | 8.60 | 细胞核Nucleus |
| TaIQM15 | TraesCS5B02G128200.1 | Chr5B: 234295411-234304552 | 480 | 53 236.89 | 6.14 | 细胞核Nucleus |
| TaIQM16 | TraesCS5B02G377900.1 | Chr5B: 555950716-555953748 | 538 | 60 186.87 | 7.57 | 细胞核Nucleus |
| TaIQM17 | TraesCS5D02G137100.1 | Chr5D: 218051693-218055671 | 480 | 53 123.76 | 6.30 | 细胞核Nucleus |
| TaIQM18 | TraesCS5D02G385200.1 | Chr5D: 454309041-454312175 | 533 | 59 832.36 | 7.17 | 细胞核Nucleus |
| TaIQM19 | TraesCS6A02G108900.1 | Chr6A: 77780102-77784144 | 485 | 53 610.33 | 8.58 | 细胞核Nucleus |
| TaIQM20 | TraesCS6B02G133900.1 | Chr6B: 130822739-130829829 | 477 | 52 647.25 | 6.44 | 细胞核Nucleus |
| TaIQM21 | TraesCS6B02G137500.1 | Chr6B: 135112248-135116198 | 483 | 53 349.14 | 8.82 | 细胞核Nucleus |
| TaIQM22 | TraesCS6D02G093400.1 | Chr6D: 58234159-58240593 | 480 | 53 062.87 | 8.04 | 细胞核Nucleus |
| TaIQM23 | TraesCS6D02G097300.1 | Chr6D: 61134482-61138500 | 484 | 53 425.06 | 8.06 | 细胞核Nucleus |
Table 2 Characteristics and subcellular localization prediction of TaIQM genes
基因 Gene | 基因号 Gene ID | 染色体定位 Chromosome localization | 氨基酸数 Number of amino acids | 相对分子量 Molecular weight/Da | 等电点 Point isoelectric | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| TaIQM1 | TraesCS1A02G125400.1 | Chr1A: 149352287-149356056 | 517 | 57 811.59 | 7.68 | 细胞核Nucleus |
| TaIQM2 | TraesCS1B02G143800.1 | Chr1B: 195555337-195560543 | 580 | 65 214.18 | 7.69 | 细胞核 Nucleus |
| TaIQM3 | TraesCS1D02G128400.1 | Chr1D: 141545585-141547817 | 424 | 47 667.67 | 9.49 | 细胞核Nucleus |
| TaIQM4 | TraesCS2A02G154900.1 | Chr2A: 102130189-102132879 | 564 | 63 624.94 | 6.47 | 细胞核Nucleus |
| TaIQM5 | TraesCS2B02G180000.1 | Chr2B: 154592307-154595019 | 572 | 64 351.52 | 6.36 | 细胞核Nucleus |
| TaIQM6 | TraesCS2D02G160200.1 | Chr2D: 103126893-103129599 | 571 | 64 331.63 | 6.26 | 细胞核Nucleus |
| TaIQM7 | TraesCS3A02G206400.1 | Chr3A: 363415357-363419189 | 556 | 62 188.59 | 9.13 | 细胞核Nucleus |
| TaIQM8 | TraesCS3B02G238500.1 | Chr3B: 373842628-373844908 | 541 | 60 758.12 | 9.07 | 细胞核Nucleus |
| TaIQM9 | TraesCS3D02G209200.1 | Chr3D: 276574646-276577885 | 530 | 59 496.51 | 8.97 | 细胞核Nucleus |
| TaIQM10 | TraesCS4A02G021000.1 | Chr4A: 14261744-14265762 | 610 | 67 879.37 | 9.60 | 细胞核Nucleus |
| TaIQM11 | TraesCS4B02G282600.2 | Chr4B: 565861371-565865127 | 669 | 74 779.20 | 9.41 | 细胞核Nucleus |
| TaIQM12 | TraesCS4D02G281600.1 | Chr4D: 452795390-452798654 | 613 | 68 128.53 | 9.67 | 细胞核Nucleus |
| TaIQM13 | TraesCS5A02G129600.1 | Chr5A: 290924878-290930090 | 461 | 51 276.55 | 6.88 | 细胞核Nucleus |
| TaIQM14 | TraesCS5A02G375700.1 | Chr5A: 573606336-573609441 | 534 | 59 693.35 | 8.60 | 细胞核Nucleus |
| TaIQM15 | TraesCS5B02G128200.1 | Chr5B: 234295411-234304552 | 480 | 53 236.89 | 6.14 | 细胞核Nucleus |
| TaIQM16 | TraesCS5B02G377900.1 | Chr5B: 555950716-555953748 | 538 | 60 186.87 | 7.57 | 细胞核Nucleus |
| TaIQM17 | TraesCS5D02G137100.1 | Chr5D: 218051693-218055671 | 480 | 53 123.76 | 6.30 | 细胞核Nucleus |
| TaIQM18 | TraesCS5D02G385200.1 | Chr5D: 454309041-454312175 | 533 | 59 832.36 | 7.17 | 细胞核Nucleus |
| TaIQM19 | TraesCS6A02G108900.1 | Chr6A: 77780102-77784144 | 485 | 53 610.33 | 8.58 | 细胞核Nucleus |
| TaIQM20 | TraesCS6B02G133900.1 | Chr6B: 130822739-130829829 | 477 | 52 647.25 | 6.44 | 细胞核Nucleus |
| TaIQM21 | TraesCS6B02G137500.1 | Chr6B: 135112248-135116198 | 483 | 53 349.14 | 8.82 | 细胞核Nucleus |
| TaIQM22 | TraesCS6D02G093400.1 | Chr6D: 58234159-58240593 | 480 | 53 062.87 | 8.04 | 细胞核Nucleus |
| TaIQM23 | TraesCS6D02G097300.1 | Chr6D: 61134482-61138500 | 484 | 53 425.06 | 8.06 | 细胞核Nucleus |

Fig. 3 Cis-acting elements analysis of TaIQM genesNote: ABRE—Cis-acting element involved in the abscisic acid responsiveness; ARE—Cis-acting regulatory element essential for the anaerobic induction; Circadian—Cis-acting regulatory element involved in circadian control; G-box—Cis-acting regulatory element involved in light responsiveness; TGACG-motif—Cis-acting regulatory element involved in the MeJA-responsiveness; GT1-motif—Light responsive element; TC-rich repeats—Cis-acting element involved in defense and stress responsiveness; G-box—Cis-acting regulatory element involved in light responsiveness; CGTCA-motif—Cis-acting regulatory element involved in the MeJA-responsiveness; CAT-box—Cis-acting regulatory element related to meristem expression; P-box—Gibberellin-responsive element; GARE-motif—Gibberellin-responsive element; TGA-element—Auxin-responsive element; ACE—Cis-acting element involved in light responsiveness; GCN4_motif—Cis-regulatory element involved in endosperm expression; TCA-element—Cis-acting element involved in salicylic acid responsiveness; TATC-box—Cis-acting element involved in gibberellin-responsiveness; MBS—MYB binding site involved in drought-inducibility; AuxRR-core—Cis-acting regulatory element involved in auxin responsiveness; LTR—Cis-acting element involved in low-temperature responsiveness.
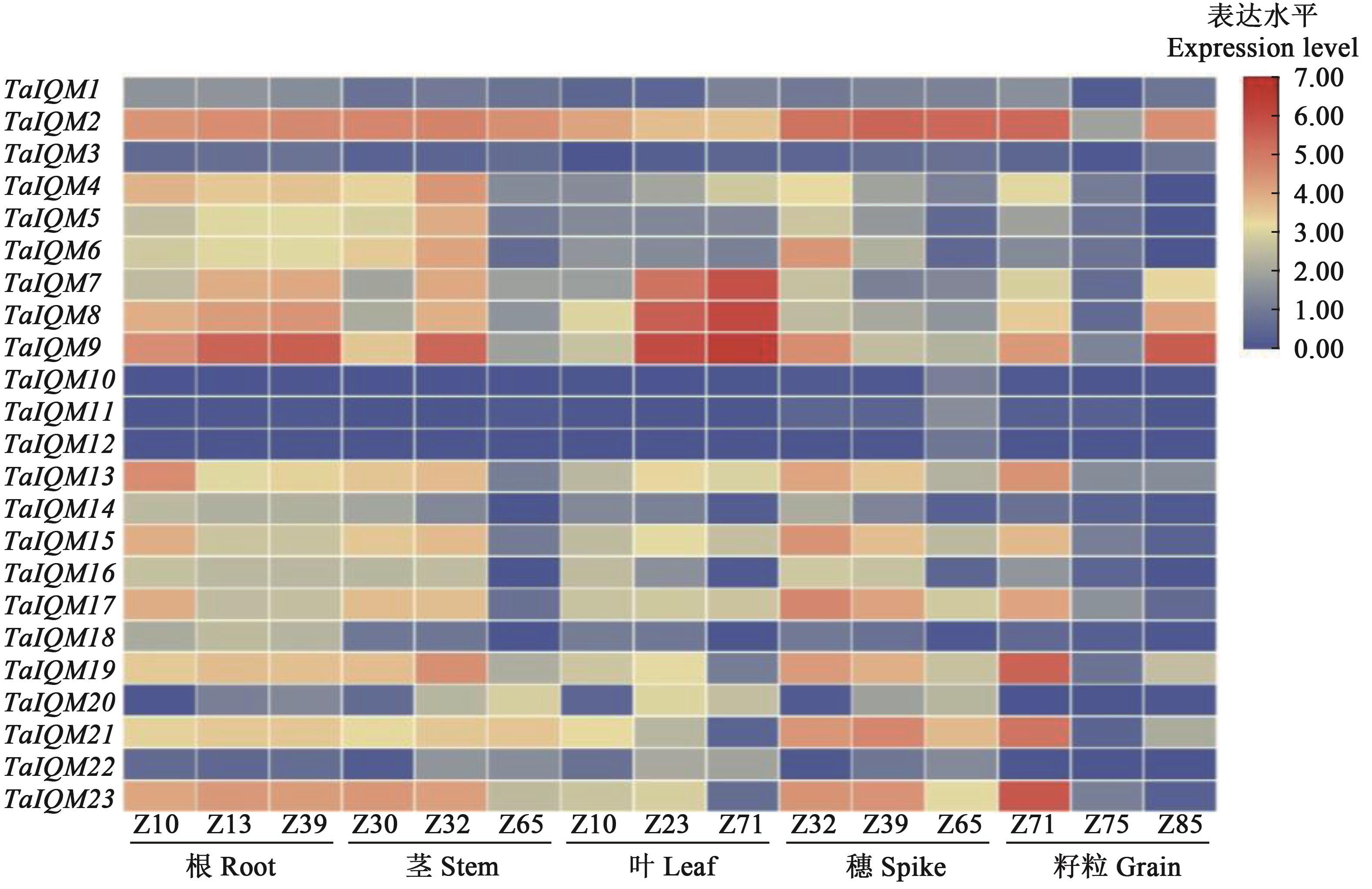
Fig. 4 Heat map of relative expression level of TaIQM genes in different tissuesNote: Red color indicates up-regulation expression, blue color indicates down-regulation expression. Z10—1 leaf period; Z13—3 leaves stage; Z23—Early tillering; Z30—Standing stage; Z32—Early jointing stage; Z39—Late jointing stage; Z65—Middle flowering; Z71—2 d after flowering; Z75—10 d after flowering; Z85—30 d after flowering.
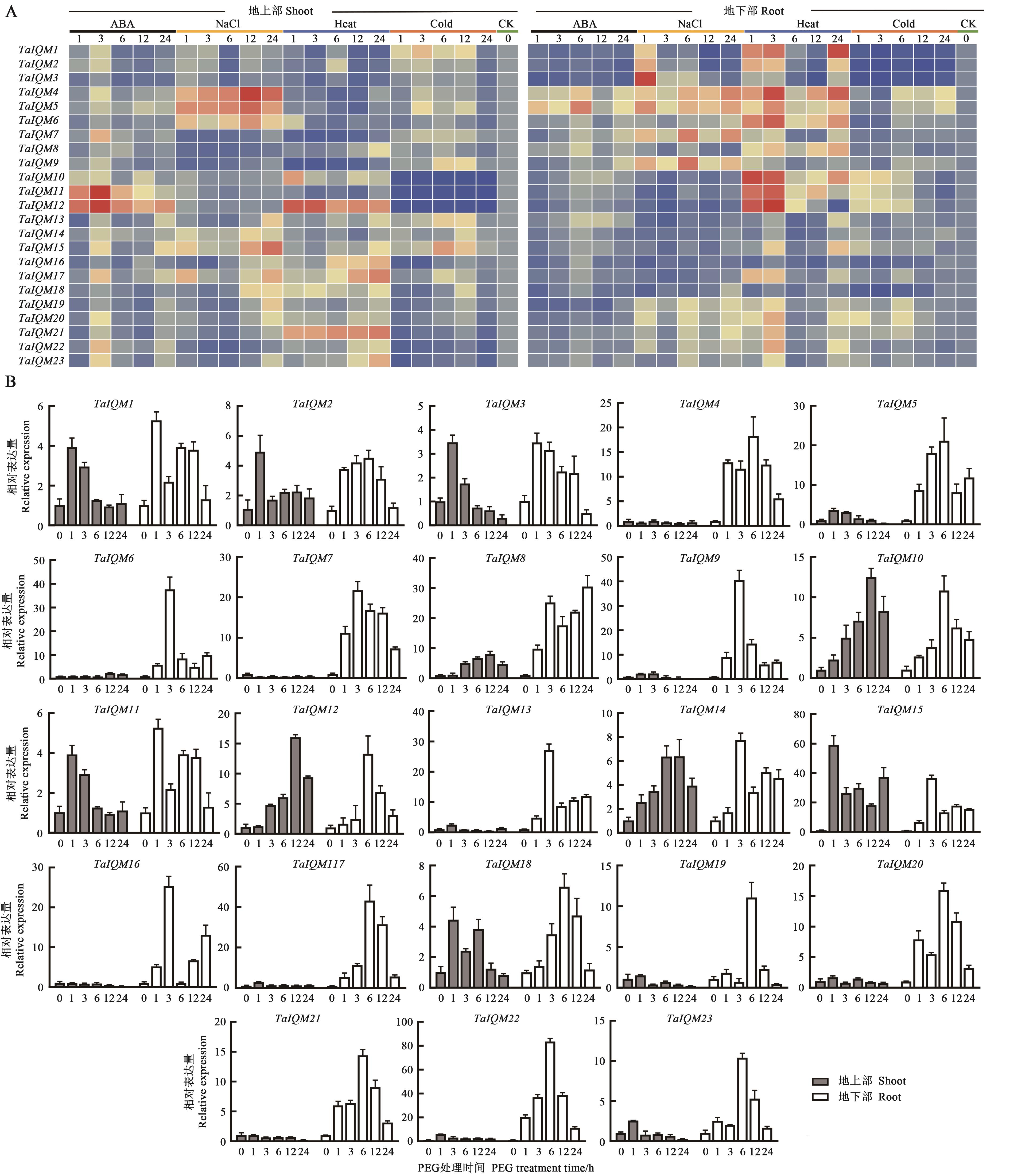
Fig. 5 Expression analysis of IQM genes in wheat under abiotic stressesA: Relative expressionunder ABA, NaCl, heat and clod; B: Relative expression under drought.Red color indicates up-regulation expression, blue color indicates down-regulation expression
| 1 | 田长恩, 周玉萍. 植物具IQ基序的钙调素结合蛋白的研究进展[J]. 植物学报, 2013,48(4):447-460. |
| TIAN C N, ZHOU Y P. Research progress in plant IQ motif-containing calmodulin-binding proteins [J]. Chin. Bull. Botany, 2013, 48(4): 447-460. | |
| 2 | ZHOU Y P, DUAN J, FUJIBE T, et al.. AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in Arabidopsis [J]. Plant Mol. Biol., 2012,79(4-5):333-346. |
| 3 | 曾后清, 张亚仙, 汪尚, 等. 植物钙/钙调素介导的信号转导系统[J]. 植物学报, 2016,51(5):705-723. |
| ZENG H Q, ZHANG Y X, WANG S, et al.. Calcium/calmodulin-mediated signal transduction systems in plants [J]. Chin. Bull. Bot., 2016,51(5):705-723. | |
| 4 | DEFALCO T A, BENDER K W, SNEDDEN W A. Breaking the code: Ca2+ sensors in plant signalling [J]. Biochem. J., 2009,425(1):27-40. |
| 5 | NG C K, MCAINSH M R, GRAY J E, et al.. Calcium-based signalling systems in guard cells [J]. New Phytol., 2001,151(1):109-120. |
| 6 | ZHOU Y P, WU J H, XIAO W H, et al.. Arabidopsis IQM 4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in Arabidopsis [J/OL]. Front. Plant Sci., 2018,9:721 [2022-06-08].. |
| 7 | FAN T, LYU T, XIE C, et al.. Genome-wide analysis of the IQM gene family in rice (Oryza sativa L.) [J/OL]. Plants, 2021,10(9):1949 [2022-06-08].. |
| 8 | PATRA N, HARIHARAN S, GAIN H, et al.. Typical but delicate Ca(2+)re: dissecting the essence of calcium signaling network as a robust response coordinator of versatile abiotic and biotic stimuli in plants [J/OL]. Front. Plant Sci., 2021,12:752246 [2022-06-08]. . |
| 9 | 罗慧婷, 吕天晓, 范甜, 等. 拟南芥IQM1互作蛋白的筛选与验证[J]. 科技视界, 2018(9):83-84. |
| LUO H T, LYU T X, FAN T, et al.. Screening and verification of the protein interacting with IQM1 in Arabidopsis [J]. Sci. Technol. Vision, 2018(9):83-84. | |
| 10 | 周玉萍, 陈琼华, 陈洁珊, 等. IQM1基因过量表达对拟南芥气孔运动及根系生长的影响[J]. 西北植物学报, 2013,33(5):904-910. |
| ZHOU Y P, CHEN Q H, CHEN J S, et al.. Overexpression of Arabidopsis IQM1 gene affects stomatal movement and root growth [J]. Acta Bot. Bor-Occid. Sin., 2013,33(5):904-910. | |
| 11 | 黄章科, 张艺能, 莫忠蓁, 等. IQ基序突变对AtIQM1的钙调素结合活性的影响[J]. 生物技术通报, 2012,12(21):128-132. |
| HUANG Z K, ZHANG Y N, MO Z Q, et al.. Effects of mutations in IQ motif of AtIQM1 on its calmodulin binding [J]. Biotechnol. Bull., 2012,12(21):128-132. | |
| 12 | 吴骏. 拟南芥IQM2参与成花调控与CaM信号关系的初步研究[D]. 广州:广州大学, 2017. |
| WU J. Preliminary study on relationship of IQM2 mediating flowering and CaM signaling in Arabidopsis [D]. Guangzhou: Guangzhou University, 2017. | |
| 13 | 徐浩, 冯奕嘉, 范甜, 等. 拟南芥IQM3基因突变减少幼苗的侧根数量和增加主根长度[J]. 植物生理学报, 2019,55(5):629-634. |
| XU H, FENG Y J, FAN T, et al.. Disruption of IQM3 reduces the number of lateral roots and increases the length of primary root in Arabidopsis seedlings [J]. Plant Physiol. J., 2019,55(5):629-634. | |
| 14 | 萧文慧, 宋俊威, 黄小玲, 等. 非生物胁迫对拟南芥IQM4基因表达的影响[J]. 科技视界, 2016(16):10-11. |
| XIAO W H, SONG J W, HUANG X L, et al.. Effect of abiotic stress on IQM4 gene expression in Arabidopsis thaliana [J]. Sci. Technol. Vision, 2016(16):10-11. | |
| 15 | 弓路平, 萧文慧, 周玉萍, 等. 拟南芥IQM5.2的克隆、表达及其生物信息学分析[J]. 生物技术通报, 2016,32(5):69-74. |
| GONG L P, XIAO W H, ZHOU Y P, et al.. Cloning,expression and bioinformatics analysis of IQM5.2 from Arabidopsis [J]. Biotechnol. Bull.,2016,32(5):69-74. | |
| 16 | 冯奕嘉, 徐浩, 范甜, 等. 拟南芥IQM6突变推迟远轴面表皮毛的发生[J]. 植物生理学报, 2019,55(6):729-735. |
| FENG Y J, XU H, FAN T, et al.. IQM6 mutantion delays initiation of abaxial trichomes in Arabidopsis [J]. Plant Physiol. J., 2019,55(6):729-735. | |
| 17 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The pfam protein families database in 2019 [J/OL]. Nucleic Acids Res., 2019,47(D1):995 [2022-06-08]. . |
| 18 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J/OL]. Nucleic Acids Res., 2002,30(1):325 [2022-06-08]. . |
| 19 | AROCHO A, CHEN B, LADANYI M, et al.. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts [J]. Diagn Mol. Pathol., 2006,15(1):56-61. |
| 20 | WU M, LI Y, CHEN D, et al.. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis) [J/OL]. Sci. Rep., 2016,6:24520 [2022-06-08]. . |
| 21 | ZHOU Y, CHEN Y, YAMAMOTO K T, et al.. Sequence and expression analysis of the Arabidopsis IQM family [J]. Acta Physiol. Plantarum., 2010,32(1):191-198. |
| 22 | ZHAO J F, ZHAO L L, ZHANG M, et al.. Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation [J/OL]. Int. J. Mol. Sci., 2017, 18(9): 1841 [2022-06-08]. . |
| 23 | 杨华杰, 周玉萍, 范甜, 等. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021,37(11):190-196. |
| YANG H J, ZHOU Y P, FAN T, et al.. Screening and identification of IQM4-interacting proteins in Arabidopsis thaliana [J]. Biotechnol. Bull., 2021,37(11):190-196. |
| [1] | Yifan LIU, Shaoshuai LIU, Rui ZANG, Yang LI, Wei LIU, Tingting LI, Danmei LIU, Dengcai LIU, Aili LI, Long MAO, Xiang WANG, Shuaifeng GENG. Analysis of Quality Traits of 168 Wheat Germplasm Resources [J]. Journal of Agricultural Science and Technology, 2025, 27(9): 44-57. |
| [2] | Caixia LYU, Yongfu LI, Huinan XIN, Na LI, Ning LAI, Qinglong GENG, Shuhuang CHEN. Effects of Slow Release Nitrogen Fertilizer on Yield of Winter Wheat and Soil Nitrate/Ammonium Nitrogen Under Drip Irrigation [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 179-186. |
| [3] | Qiang ZHU, Zongxian CHE, Heng CUI, Jiudong ZHANG, Xingguo BAO. Effect of Green Manure Replacing Nitrogen Fertilizer on Greenhouse Gases in Wheat Fields [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 182-189. |
| [4] | Yi HU, Jie GONG, Wei ZHAO, Rong CHENG, Zhongyu LIU, Shiqing GAO, Yazhen YANG. Identification of PHY Gene Family in Wheat (Triticum aestivum L.)and Their Expression Analysis Under Heat Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 30-43. |
| [5] | Sile HU, Yulong BAO, Tubuxinbayaer, Jifeng TAO, Enliang GUO. Chlorophyll Content Inversion of Spring Wheat Based on Unmanned Aerial Vehicle Hyperspectral and Integrated Learning [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 93-103. |
| [6] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [7] | Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35. |
| [8] | Yixin CHEN, Xiubo YANG, Shijun TIAN, Cong WANG, Zhiying BAI, Cundong LI, Ke ZHANG. Response of GhCOMT28 to Drought Stress in Gossypium hirsutum [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 45-56. |
| [9] | Zhenyu XUE, Kangkang ZHANG, Yuanyuan ZHANG, Qiangqiang YAN, Lirong YAO, Hong ZHANG, Yaxiong MENG, Erjing SI, Baochun LI, Xiaole MA, Huajun WANG, Juncheng WANG. Screening and Functional Gene Detection of High-quality and Drought-resistant Wheat Germplasms [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 35-49. |
| [10] | Xianyin SUN, Qiuhuan MU, Yong MI, Guangde LYU, Xiaolei QI, Yingying SUN, Xundong YIN, Ruixia WANG, Ke WU, Zhaoguo QIAN, Yan ZHAO, Minggang GAO. Classification and Evaluation of New Wheat Lines Based on GT Biplot [J]. Journal of Agricultural Science and Technology, 2024, 26(7): 14-24. |
| [11] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [12] | Yangyang DU, Yuanyuan BAO, Xiangyu LIU, Xinyong ZHANG. Effects of Tartary Buckwheat Rotation on Enzyme Activities and Microorganisms in Rhizosphere Soil of Cultivated Potato in Yunnan Province [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 192-200. |
| [13] | Gang ZHAO, Shuying WANG, Shangzhong LI, Jianjun ZHANG, Yi DANG, Lei WANG, Xingmao LI, Wanli CHENG, Gang ZHOU, Shengli NI, Tinglu FAN. Effects of Precipitation on Yield and Water Consumption of Winter Wheat in Loess Plateau in Recent 40 Years [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 164-173. |
| [14] | Hong ZHANG, Weiguo LI, Xiaodong ZHANG, Bihui LU, Chengcheng ZHANG, Wei LI, Tinghuai MA. Extraction of Winter Wheat Planting Area Based on Fusion Features of HJ-1 and GF-1 Image [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 109-119. |
| [15] | Jingyun ZHANG, Feng GUAN, Bo SHI, Xinjian WAN. Effects of Wheat Root Exudates on Bitter Gourd Seeding Growth and Soil Environment [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 181-190. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号