




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (8): 151-162.DOI: 10.13304/j.nykjdb.2023.0338
张倩1,2( ), 门丽娜1(
), 门丽娜1( ), 李一然1, 刘巧1, 胡晓雯1, 张宇宏2(
), 李一然1, 刘巧1, 胡晓雯1, 张宇宏2( ), 张志伟1(
), 张志伟1( ), 张伟2
), 张伟2
出版日期:2024-08-15
发布日期:2024-08-12
通讯作者:
张宇宏,张志伟
基金资助:
Qian ZHANG1,2( ), Lina MEN1(
), Lina MEN1( ), Yiran LI1, Qiao LIU1, Angie DENG3, Xiaowen HU1, Yuhong ZHANG2(
), Yiran LI1, Qiao LIU1, Angie DENG3, Xiaowen HU1, Yuhong ZHANG2( ), Zhiwei ZHANG1(
), Zhiwei ZHANG1( ), Wei ZHANG2
), Wei ZHANG2
Online:2024-08-15
Published:2024-08-12
Contact:
Yuhong ZHANG,Zhiwei ZHANG
About author:ZHANG Qian E-mail: 16635047582@163.com摘要:
桃蛀果蛾是严重隐蔽危害的蛀果害虫,亟待开发环境友好型防治方法,昆虫行为调控技术被认为是新型绿色的害虫防控手段,明晰嗅觉识别机制是开发行为调控剂的基础。基于桃蛀果蛾触角转录组得到242个嗅觉基因,基于序列比对和近缘物种相似基因的功能背景,选取了13个嗅觉基因,采用RT-qPCR进一步分析其潜在识别功能。结果表明,13个嗅觉基因中有6个基因在雌雄虫不同发育时期的表达水平存在显著差异( P<0.05),推测这6个基因与成虫期嗅觉识别行为相关;根据雌雄成虫初羽化、成熟未交尾和交尾后基因表达水平的差异推测, OBP11可能在识别性信息素和交配行为中发挥重要作用,OR45可能与雌虫识别寄主植物和产卵过程相关,IR5可能与交配和产卵行为相关。以上研究结果为解析桃蛀果蛾嗅觉识别机制奠定了基础,同时也为开发新型昆虫行为调控剂、构建环境友好的害虫治理策略提供了思路。
中图分类号:
张倩, 门丽娜, 李一然, 刘巧, 胡晓雯, 张宇宏, 张志伟, 张伟. 桃蛀果蛾雌雄虫不同发育时期嗅觉基因的表达水平差异[J]. 中国农业科技导报, 2024, 26(8): 151-162.
Qian ZHANG, Lina MEN, Yiran LI, Qiao LIU, Angie DENG, Xiaowen HU, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Differential Expression Paradigm of Chemoreceptor Genes Between Males and Females at Different Developmental Stages of Carposina sasakii Matsumura[J]. Journal of Agricultural Science and Technology, 2024, 26(8): 151-162.
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| obp11 | AAAATGCTGTCCCTCCTGCC | GGCATAAGTCAGAGCCACCT |
| pbp2 | TGGATCCTGATGGCAAGCTG | GCCATCTTTGAAGCAGCGAG |
| pbp3 | AAACGCTGTCTCCTTGTCGG | TCCGACCTCCTTGCTTACCA |
| csp8 | GTCCGGCTCTGCAAGATGTA | AGAGGACGCCACAAATCTCG |
| or2 | CGCCTCTTCTGAACCGTCAT | CACGCTCACTCTACTCGCAT |
| or5 | GTCTCGCAGCTCCCATACAA | CATTACGGCCTCTACCGTGG |
| or14 | TGAGCATAAGATCCCGACGC | TGCTTTTTCTCGTTCACCTGT |
| or21 | AGCAGATGGAGAATCCAGCG | TCGCAACCATTCCCGTTGTA |
| or45 | GGAACAAACGCAGACCAACA | GGGATGAAAGGTGGCAGGAG |
| or48 | AAGCAGAGCAGTAAAGCGGA | TATGCCAGCGCCAAGAGATT |
| ir1 | ACGCTTTGTTGGAGTGACCA | ATATTCGGGCACGTCAGGTC |
| ir5 | CGGCAGAAAGGGAAGAGGTT | AAAGCCGGTGAGGACTAACG |
| ir7 | ACCCTGTTAGCCGCATCAAA | CCCAAGGGTCACAATCGACA |
Table 1 Primers used for RT?qPCR
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| obp11 | AAAATGCTGTCCCTCCTGCC | GGCATAAGTCAGAGCCACCT |
| pbp2 | TGGATCCTGATGGCAAGCTG | GCCATCTTTGAAGCAGCGAG |
| pbp3 | AAACGCTGTCTCCTTGTCGG | TCCGACCTCCTTGCTTACCA |
| csp8 | GTCCGGCTCTGCAAGATGTA | AGAGGACGCCACAAATCTCG |
| or2 | CGCCTCTTCTGAACCGTCAT | CACGCTCACTCTACTCGCAT |
| or5 | GTCTCGCAGCTCCCATACAA | CATTACGGCCTCTACCGTGG |
| or14 | TGAGCATAAGATCCCGACGC | TGCTTTTTCTCGTTCACCTGT |
| or21 | AGCAGATGGAGAATCCAGCG | TCGCAACCATTCCCGTTGTA |
| or45 | GGAACAAACGCAGACCAACA | GGGATGAAAGGTGGCAGGAG |
| or48 | AAGCAGAGCAGTAAAGCGGA | TATGCCAGCGCCAAGAGATT |
| ir1 | ACGCTTTGTTGGAGTGACCA | ATATTCGGGCACGTCAGGTC |
| ir5 | CGGCAGAAAGGGAAGAGGTT | AAAGCCGGTGAGGACTAACG |
| ir7 | ACCCTGTTAGCCGCATCAAA | CCCAAGGGTCACAATCGACA |
| Sample | ReadSum | BaseSum | GC/% | N/% | Q20/% | Cycle Q20/% | Q30/% |
|---|---|---|---|---|---|---|---|
| 1M | 34 008 437 | 8 564 069 432 | 45.38 | 0.00 | 91.76 | 99.60 | 85.84 |
| 2M | 31 812 028 | 8 012 439 316 | 44.85 | 0.00 | 91.98 | 99.60 | 86.20 |
| 3M | 31 090 051 | 7 829 911 646 | 45.09 | 0.00 | 91.84 | 99.60 | 86.03 |
| 1F | 31 277 367 | 7 875 117 126 | 45.14 | 0.00 | 91.85 | 99.60 | 85.99 |
| 2F | 25 584 959 | 6 443 563 720 | 45.03 | 0.00 | 92.56 | 99.60 | 86.86 |
| 3F | 25 864 927 | 6 513 965 846 | 44.78 | 0.00 | 91.67 | 99.60 | 85.55 |
Table 2 Statistical table of C. sasakii sequencing data
| Sample | ReadSum | BaseSum | GC/% | N/% | Q20/% | Cycle Q20/% | Q30/% |
|---|---|---|---|---|---|---|---|
| 1M | 34 008 437 | 8 564 069 432 | 45.38 | 0.00 | 91.76 | 99.60 | 85.84 |
| 2M | 31 812 028 | 8 012 439 316 | 44.85 | 0.00 | 91.98 | 99.60 | 86.20 |
| 3M | 31 090 051 | 7 829 911 646 | 45.09 | 0.00 | 91.84 | 99.60 | 86.03 |
| 1F | 31 277 367 | 7 875 117 126 | 45.14 | 0.00 | 91.85 | 99.60 | 85.99 |
| 2F | 25 584 959 | 6 443 563 720 | 45.03 | 0.00 | 92.56 | 99.60 | 86.86 |
| 3F | 25 864 927 | 6 513 965 846 | 44.78 | 0.00 | 91.67 | 99.60 | 85.55 |
| Length range/bp | Unigenes | Percentage of all unigenes/% |
|---|---|---|
| 200~300 | 36 325 | 42.48 |
| 301~500 | 22 047 | 25.79 |
| 501~1 000 | 12 611 | 14.75 |
| 1 001~2 000 | 7 243 | 8.47 |
| 2 000+ | 7 275 | 8.51 |
| Total number | 85 501 | |
| Total length/bp | 62 718 690 | |
| N50 length/bp | 1 515 | |
| Mean length/bp | 733.543 4 |
Table 3 Summary of data related to unigene
| Length range/bp | Unigenes | Percentage of all unigenes/% |
|---|---|---|
| 200~300 | 36 325 | 42.48 |
| 301~500 | 22 047 | 25.79 |
| 501~1 000 | 12 611 | 14.75 |
| 1 001~2 000 | 7 243 | 8.47 |
| 2 000+ | 7 275 | 8.51 |
| Total number | 85 501 | |
| Total length/bp | 62 718 690 | |
| N50 length/bp | 1 515 | |
| Mean length/bp | 733.543 4 |
| Annotated database | Unigenes | Percentage/% |
|---|---|---|
| COG annotation | 5 159 | 29.55 |
| GO annotation | 8 968 | 51.37 |
| KEGG annotation | 6 217 | 35.61 |
| KOG annotation | 9 969 | 57.11 |
| Pfam annotation | 12 127 | 69.47 |
| Swissprot_annotation | 8 825 | 50.55 |
| eggNOG annotation | 15 553 | 89.09 |
| Nr annotation | 16 753 | 95.97 |
| All annotated | 17 457 | 100.00 |
Table 4 Summary of database annotations
| Annotated database | Unigenes | Percentage/% |
|---|---|---|
| COG annotation | 5 159 | 29.55 |
| GO annotation | 8 968 | 51.37 |
| KEGG annotation | 6 217 | 35.61 |
| KOG annotation | 9 969 | 57.11 |
| Pfam annotation | 12 127 | 69.47 |
| Swissprot_annotation | 8 825 | 50.55 |
| eggNOG annotation | 15 553 | 89.09 |
| Nr annotation | 16 753 | 95.97 |
| All annotated | 17 457 | 100.00 |
| Olfactory gene | Stages team | Total | |||||
|---|---|---|---|---|---|---|---|
| 1M | 2M | 3M | 1F | 2F | 3F | ||
| obp | 38 | 38 | 34 | 39 | 39 | 37 | 40 |
| pbp | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| csp | 15 | 14 | 15 | 14 | 13 | 16 | 16 |
| or | 67 | 68 | 63 | 61 | 64 | 62 | 74 |
| orco | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ir | 11 | 12 | 10 | 11 | 14 | 12 | 14 |
| gr | 71 | 79 | 68 | 67 | 68 | 66 | 88 |
| ode | 6 | 4 | 5 | 4 | 5 | 5 | 6 |
Table 5 Differentially expressed genes related to olfaction in 3 stages of C. sasakii
| Olfactory gene | Stages team | Total | |||||
|---|---|---|---|---|---|---|---|
| 1M | 2M | 3M | 1F | 2F | 3F | ||
| obp | 38 | 38 | 34 | 39 | 39 | 37 | 40 |
| pbp | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| csp | 15 | 14 | 15 | 14 | 13 | 16 | 16 |
| or | 67 | 68 | 63 | 61 | 64 | 62 | 74 |
| orco | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ir | 11 | 12 | 10 | 11 | 14 | 12 | 14 |
| gr | 71 | 79 | 68 | 67 | 68 | 66 | 88 |
| ode | 6 | 4 | 5 | 4 | 5 | 5 | 6 |
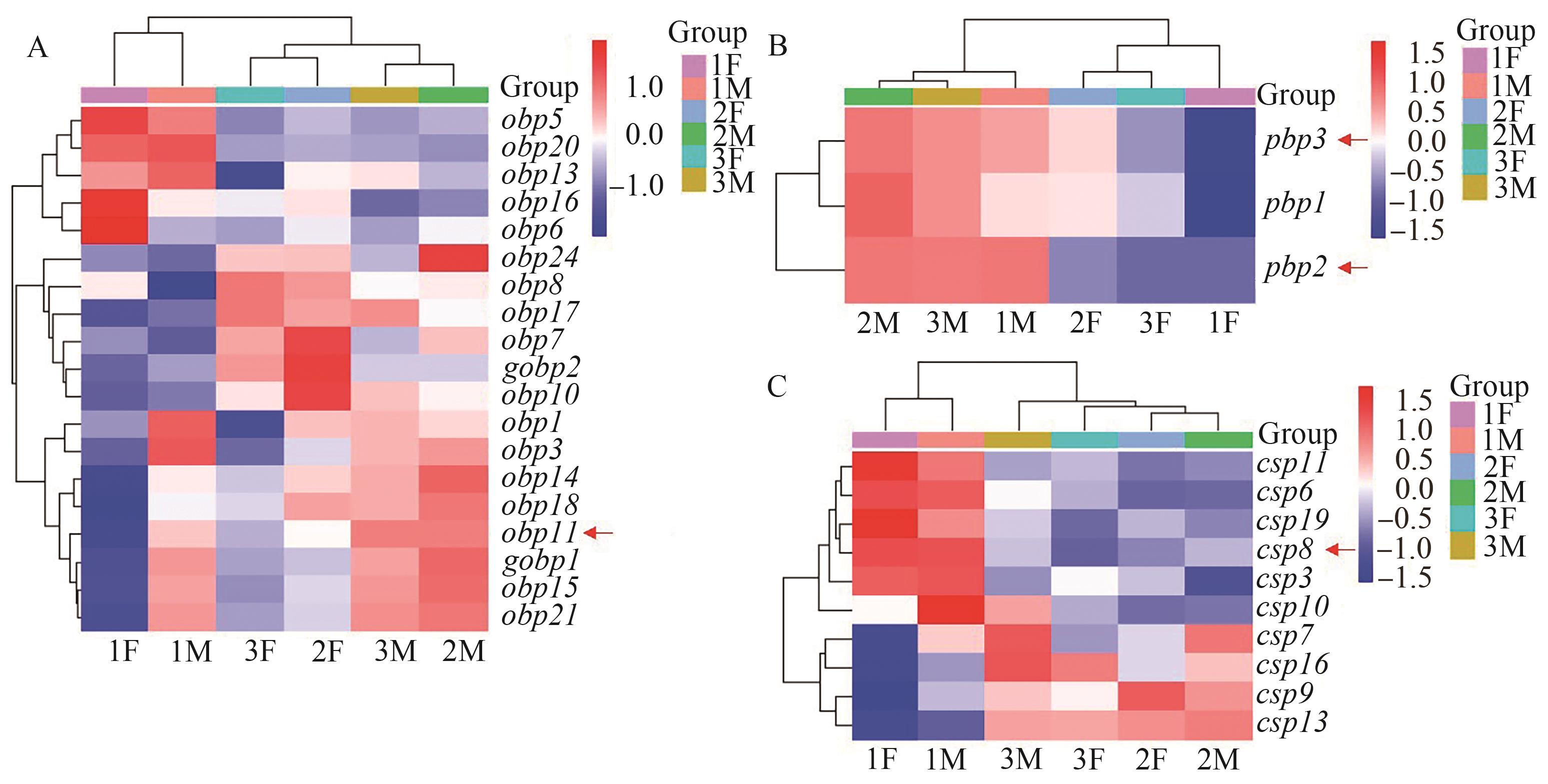
Fig. 2 Differential expression of semiochemical binding genesA: Differential expression of obps genes; B: Differential expression of pbps genes; C: Differential expression of csps genes. Red arrows show the interested differentially expressed genes
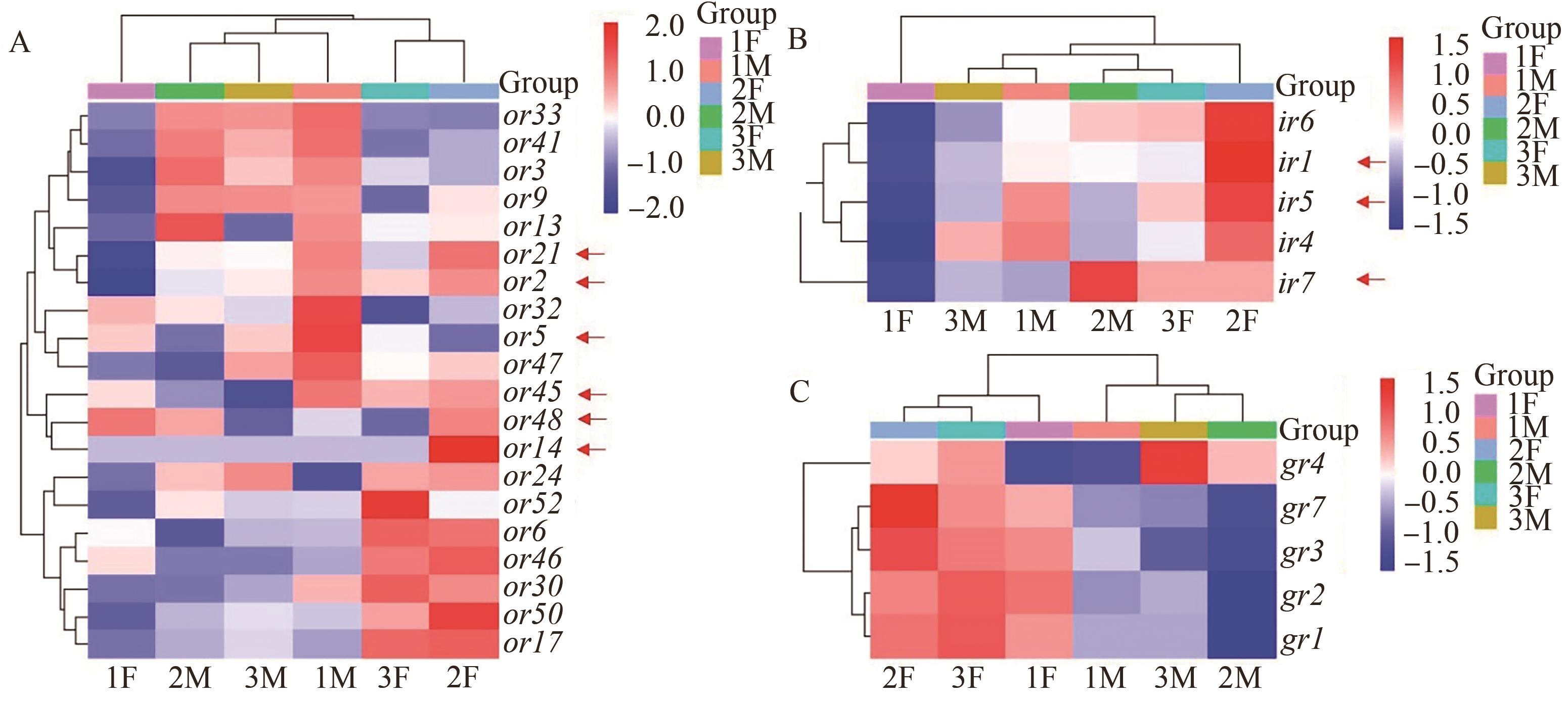
Fig. 3 Differential expression of semiochemical receptor genesA: Differential expression of ors genes; B: Differential expression of irs genes; C: Differential expression of grs genes. Red arrows show the interested differentially expressed genes
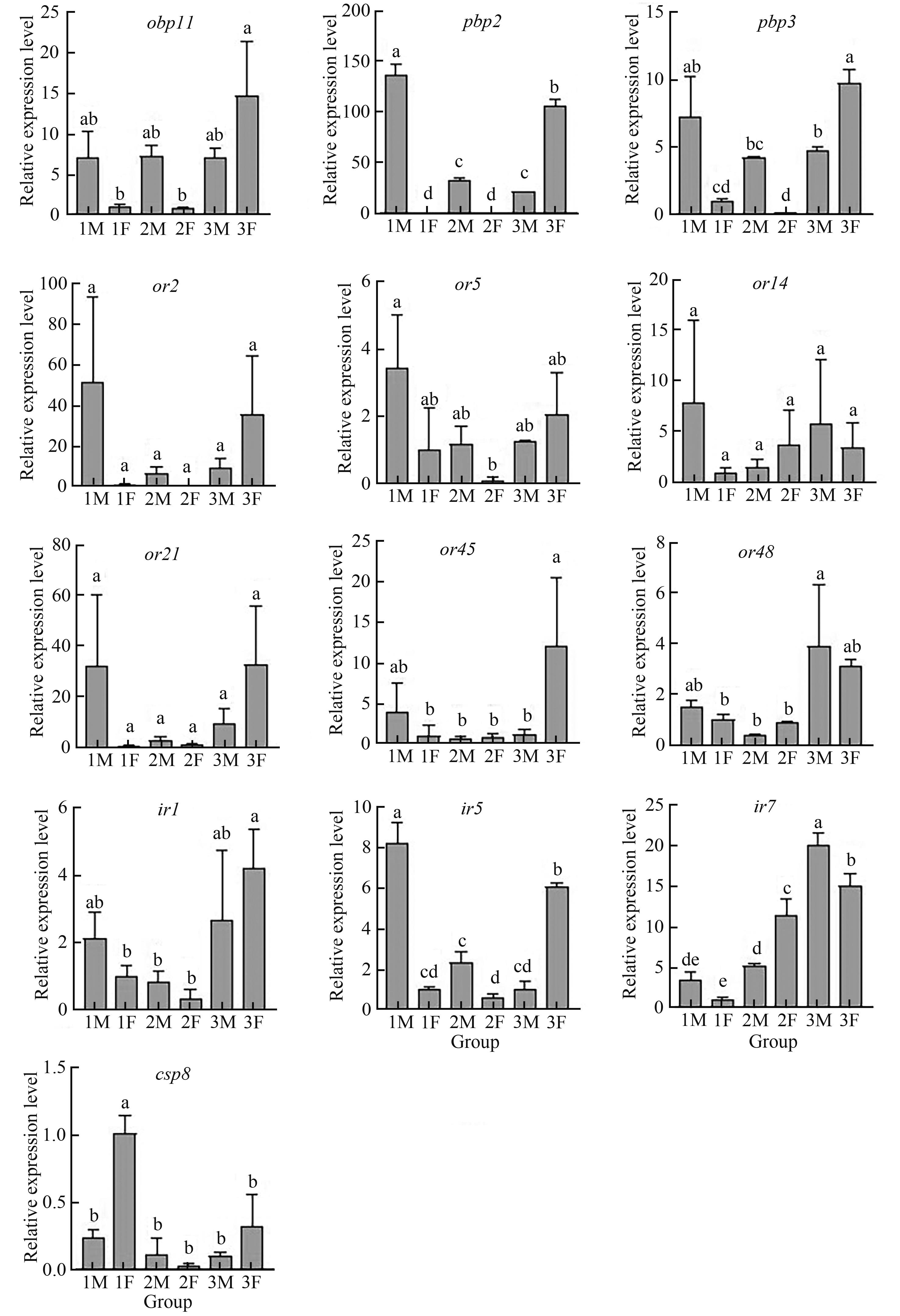
Fig. 4 Different sex and developmental stage expression profiles of 15 olfactory?related genes by RT?qPCRNote:Different lowercase letters indicate significant differences between different treatments at P<0.05 level.
| 1 | LIU B, ZHAN G P, REN L L, et al.. Toxicity of pure phosphine to Carposina sasakii Matsumura (Lepidoptera: Carposinadae) [J]. Plant Prot., 2016, 42( 6): 191- 196. |
| 2 | KIM D S, LEE J H, YIEM M S. Temperature-dependent development of Carposina sasakii (Lepidoptera: Carposinidae) and its stage emergence models [J]. Environ. Entomol., 2001, 30( 2): 298- 305. |
| 3 | HUA B Z, ZENG X H, ZHANG H. Influences of apple maturity on the development and diapause of Carposina sasakii Matsumura [J]. J. Northwest A&F Univ., 1996, 24( 6): 42- 45. |
| 4 | FENG D D, XUE Q Q, MEN L N, et al.. Demography and mass-rearing of Carposina sasakii Matsumura (Lepidoptera: Carposinidae) reared on golden delicious and red fuji apples in the laboratory [J]. J. Asia-Pac. Entomol., 2020, 23( 4): 1194- 1201. |
| 5 | ZHANG Z W, LI X W, XUE Y H, et al.. Increased trapping efficiency for the peach fruit moth Carposina sasakii (Matsumura) with synthetic sex pheromone [J]. Agric. For. Entomol., 2017, 19( 4): 424- 432. |
| 6 | YANG H B, DONG J F, SUN Y L, et al.. Antennal transcriptome analysis and expression profiles of putative chemosensory soluble proteins in Histia rhodope Cramer (Lepidoptera: Zygaenidae) [J/OL]. CBP, Part D: Genomics Proteomics, 2020, 33: 100654 [ 2023-03-25]. . |
| 7 | HU P, WANG J Z, CUI M M, et al.. Antennal transcriptome analysis of the asian longhorned beetle Anoplophora glabripennis [J/OL]. Sci. Rep., 2016, 6: 26652 [ 2023-03-25]. . |
| 8 | CHOO Y M, XU P X, HWANG J K, et al.. Reverse chemical ecology approach for the identification of an oviposition attractant for culex quinquefasciatus [J]. Proc. Natl. Acad. Sci. USA, 2018, 115: 714- 719. |
| 9 | KEPCHIA D, MOLIVER S, CHOHAN K, et al.. Inhibition of insect olfactory behavior by an airborne antagonist of the insect odorant receptor co-receptor subunit [J/OL]. PLoS One, 2017, 12( 8):e 0183009 [ 2023-03-25]. . |
| 10 | CAO S, SUN D D, LIU Y, et al.. Mutagenesis of odorant coreceptor orco reveals the distinct role of olfaction between sexes in Spodoptera frugiperda [J/OL]. J. Integr. Agric., 2022, 11: 004 [ 2023-03-25]. . |
| 11 | GRABHERR M G, HAAS B J, YASSOUR M, et al.. Full length transcriptome assembly from RNA seq data without a reference genome [J]. Nat. Biotechnol., 2011, 29: 644- 652. |
| 12 | TRAPNELL C, WILLIAMS B A, PERTEA G, et al.. Transcript assembly and quantification by RNA seq reveals unannotated transcripts and isoform switching during cell differentiation [J]. Nat. Biotechnol., 2010, 28( 5): 511- 515. |
| 13 | LI B, DEWEY C N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome [J/OL]. BMC Bioinf., 2011, 12: 323 [ 2023-03-25]. . |
| 14 | WANG E H, WANG X Q, LI H T, et al.. Effective extraction of total RNA from Carposina sasakii and the clone of the gene β⁃ actin [J]. J. Shenyang Agric. Univ., 2014, 45( 4): 403- 407. |
| 15 | SCHMITTGEN T D, LIVAK K J. Analyzing real-time PCR data by the comparative C(T) method [J]. Nat. Protocol, 2008, 3( 6): 1101- 1108. |
| 16 | LAN X N, XIANG S S, ZHU H. Research progress of the types and functions of insect antennal sensilla [J]. J. Environ. Entomol., 2023, 45( 5): 1197- 1216.. |
| 17 | CHENG J, WANG C Y, LYU Z H, et al.. Candidate olfactory genes identified in Heortia vitessoides (Lepidoptera: Crambidae) by antennal transcriptome analysis [J]. CBP, Part D: Genomics Proteomics, 2019, 29: 117- 130. |
| 18 | BRITO N F, MOREIRA M F, MELO A C. A look inside odorant-binding proteins in insect chemoreception [J]. J. Insect Physiol., 2016, 95: 51- 65. |
| 19 | LEAL W S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes [J]. Annu. Rev. Entomol., 2013, 58: 373- 391. |
| 20 | LIU N Y, ZHANG T, YE Z F, et al.. Identification and characterization of candidate chemosensory gene families from Spodoptera exigua developmental transcriptomes [J]. Int. J. Biol. Sci., 2015, 11( 9): 1036- 1048. |
| 21 | TIAN Z Q, SUN L N, LI Y Y, et al.. Antennal transcriptome analysis of the chemosensory gene families in Carposina sasakii (Lepidoptera: Carposinidae) [J/OL]. BMC Genomics, 2018, 19: 544 [ 2023-03-25]. . |
| 22 | ZHOU S S, SUN Z, MA W H, et al.. De novo analysis of the Nilaparvata lugens (Stål) antenna transcriptome and expression patterns of olfactory genes [J]. CBP, Part D: Genomics Proteomics, 2014, 9: 31- 39. |
| 23 | ANTONY B, JOHNY J, ALDOSARI S A. Silencing the odorant binding protein RferOBP1768 reduces the strong preference of palm weevil for the major aggregation pheromone compound ferrugineol [J/OL]. Front. Physiol., 2018, 9: 252 [ 2023-03-25]. . |
| 24 | ZHANG Y Y, GUO J M, WEI Z Q, et al.. Identification and sex expression profiles of olfactory-related genes in Mythimna loreyi based on antennal transcriptome analysis [J/OL]. J. Insect Sci., 2022, 25( 3): 101934 [ 2023-03-25]. . |
| 25 | LIU P J, ZHANG X F, MENG R J, et al.. Identification of chemosensory genes from the antennal transcriptome of Semiothisa cinerearia [J/OL]. PLoS One, 2020, 15( 8):e 0237134 [ 2023-03-25]. . |
| 26 | PELOSI P, IOVINELLA I, FELICIOLI A, et al.. Soluble proteins of chemical communication: an overview across arthropods [J/OL]. Front. Physiol., 2014, 5: 320 [ 2023-03-25]. . |
| 27 | PELOSI P, IOVINELLA I, ZHU J, et al.. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects [J]. Biol. Rev. Cambridge Philos. Soc., 2018, 93: 184- 200. |
| 28 | ZHANG Y, FENG K, MEI R L, et al.. Analysis of the antennal transcriptome and identification of tissue-specific expression of olfactory-related genes in Micromelalopha troglodyta (Lepidoptera: Notodontidae) [J/OL]. J. Insect Sci., 2022, 22( 5): 8 [ 2023-03-25]. . |
| 29 | HA T S, SMITH D P. Odorant and pheromone receptors in insects [J/OL]. Front. Cell. Neurosci., 2009, 3: 10 [ 2023-03-25]. . |
| 30 | WICHER D, MIAZZI F. Functional properties of insect olfactory receptors: ionotropic receptors and odorant receptors [J]. J. Cell Tissue Res., 2021, 383( 1SI): 7- 19. |
| 31 | HU P, TAO J, CUI M M, et al.. Antennal transcriptome analysis and expression profiles of odorant binding proteins in Eogystia hippophaecolus (Lepidoptera: Cossidae) [J/OL]. BMC Genomics, 2016, 17: 651 [ 2023-03-25]. . |
| 32 | NIE H, XU S P, XIE C Q, et al.. Comparative transcriptome analysis of Apis mellifera antennae of workers performing different tasks [J]. Mol. Genet. Genomics, 2018, 293: 237- 248. |
| 33 | LIU Y P, DU L X, ZHU Y, et al.. Identification and sex-biased profiles of candidate olfactory genes in the antennal transcriptome of the parasitoid wasp Cotesia vestalis [J/OL]. CBP, Part D: Genomics Proteomics, 2020, 34: 100657 [ 2023-03-25]. . |
| 34 | CLAUDIANOS C, LIM J, YOUNG M, et al.. Odor memories regulate olfactory receptor expression in the sensory periphery [J]. Eur. J. Neurosci., 2014, 39: 1642- 1654. |
| 35 | WANG S N, PENG Y, LU Z Y, et al.. Identification and expression analysis of putative chemosensory receptor genes in microplitis mediator by antennal transcriptome Screening [J]. Int. J. Biol. Sci., 2015, 11( 7): 737- 751. |
| 36 | XIA Q Y, ZHOU Z Y, LU C, et al.. A draft sequence for the genome of the domesticated silkworm ( Bombyx mori) [J]. Science, 2004, 306( 5703): 1937- 1940. |
| 37 | ZHAO H T, GAO P F, ZHANG G X, et al.. Expression and localization analysis of the odorant receptor gene Orco in drones antennae of Apis cerana cerana [J]. Sci. Agric. Sin., 2015, 48( 4): 796- 803. |
| 38 | SAXENA K N, GOYAL S. Host-plant relations of the citrus butterfly Papilio demoleus L.: Orientational and ovipositional responses [J]. Entomol. Exp. Appl., 1978, 24( 1): 1- 10. |
| 39 | RYTZ R, CROSET V, BENTON R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond Insect [J]. Insect Biochem. Mol. Biol., 2013, 43( 9): 888- 897. |
| 40 | HU J, WANG X Y, TAN L S, et al.. Identification of chemosensory genes, including candidate pheromone receptors, in Phauda flammans (Walker) (Lepidoptera: Phaudidae) through transcriptomic analyses [J/OL]. Front. Physiol., 2022, 13: 907694 [ 2023-03-25]. . |
| 41 | POUDEL S, KIM Y, KIM Y T, et al.. Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster [J]. Insect Biochem. Mol. Biol., 2015, 66: 110- 118. |
| 42 | SCOTT K, BRADY R J, CRAVCHIK A, et al.. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila [J]. Cell, 2001, 104: 661- 673. |
| 43 | CHANG X Q, NIE X P, ZHANG Z, et al.. De novo analysis of the oriental armyworm Mythimna separata antennal transcriptome and expression patterns of odorant-binding proteins [J]. CBP, Part D: Genomics Proteomics, 2017, 22: 120- 130. |
| [1] | 史硕, 冯宇, 李亮, 孟瑞, 章延泽, 杨秀荣. 印度梨形孢介导小麦抗纹枯病的转录组分析及关键基因筛选[J]. 中国农业科技导报, 2025, 27(5): 133-145. |
| [2] | 王昕昕, 吴子薇, 方婷, 牟巧, 王芳, 赵晗, 杜志强, 杨彩侠. 维生素C调节小鼠TM4支持细胞的功能和基因表达[J]. 中国农业科技导报, 2025, 27(5): 81-89. |
| [3] | 鲁一薇, 夏雪岩, 赵宇, 崔纪菡, 刘猛, 黄玫红, 褚程, 刘建军, 李顺国. 缺钾胁迫下谷子转录组分析及相关基因挖掘[J]. 中国农业科技导报, 2024, 26(6): 30-44. |
| [4] | 徐佳睿, 王逸茹, 赵绍赓, 李坤, 郑军. 玉米木质素合成途径基因ZmCCoAOMT1功能研究及转录组分析[J]. 中国农业科技导报, 2024, 26(5): 30-43. |
| [5] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| [6] | 赵展, 王晓婷, 张黎凤, 赵津禾, 于玉红, 李军华, 吴占清. 西瓜对低氮胁迫响应的转录组分析[J]. 中国农业科技导报, 2024, 26(12): 30-38. |
| [7] | 陈永孜, 王化, 王维轩. 空间转录组学的发展及其应用进展[J]. 中国农业科技导报, 2024, 26(11): 23-31. |
| [8] | 夏雪岩, 崔纪菡, 黄玫红, 郭帅, 刘猛, 赵宇, 鲁一薇, 赵文庆, 王京新, 李顺国. 谷子苗期氮高效转录组分析与基因挖掘[J]. 中国农业科技导报, 2024, 26(10): 41-57. |
| [9] | 冀薇, 樊莹, 黄家兴, 杨慧鹏, 徐进, 李小英, 郭岳琴, 吴跃国, 李继莲, 姚军. 不同授粉强度对蓝莓柱头响应机制的转录组分析[J]. 中国农业科技导报, 2024, 26(10): 71-82. |
| [10] | 徐皖菁, 彭芳, 赵豆豆, 罗姣姣, 陶珊, 廖海浪, 毛常清, 吴宇, 朱秀, 徐正君, 张超. 基于转录组和代谢组解析川芎对镉胁迫的响应机制[J]. 中国农业科技导报, 2024, 26(10): 98-109. |
| [11] | 李相吴, 刘自扬, 徐玉俊, 祝建波, 吴燕民. 真菌诱导子调控紫草素合成的分子机制探究[J]. 中国农业科技导报, 2024, 26(1): 78-88. |
| [12] | 王潇然, 李笑语, 孙慧, 于海东, 石永春. 硼胁迫下烟草叶片转录组分析[J]. 中国农业科技导报, 2023, 25(8): 53-64. |
| [13] | 贾晶莹, 李雅辉, 伏兵哲, 马云, 蔡小艳. 苜蓿miRs表达谱分析及跨界潜力miRs初步筛选[J]. 中国农业科技导报, 2023, 25(7): 43-53. |
| [14] | 王云胜, 陈银翠, 程在, 张锦, 张传博. 过表达veA基因对冠突散囊菌次级代谢的影响[J]. 中国农业科技导报, 2023, 25(7): 77-86. |
| [15] | 马蓝, 彭晴, 徐小轻, 杨硕, 张宇微, 田丹丹, 施琳波, 石波, 乔宇. 大肠杆菌O157∶H7生物被膜状态下基因表达分析[J]. 中国农业科技导报, 2023, 25(6): 71-88. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||