




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (1): 78-88.DOI: 10.13304/j.nykjdb.2022.0622
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Xiangwu LI1,2( ), Ziyang LIU2,3, Yujun XU2, Jianbo ZHU1(
), Ziyang LIU2,3, Yujun XU2, Jianbo ZHU1( ), Yanmin WU2,3(
), Yanmin WU2,3( )
)
Received:2022-07-23
Accepted:2022-09-28
Online:2024-01-15
Published:2024-01-08
Contact:
Jianbo ZHU,Yanmin WU
李相吴1,2( ), 刘自扬2,3, 徐玉俊2, 祝建波1(
), 刘自扬2,3, 徐玉俊2, 祝建波1( ), 吴燕民2,3(
), 吴燕民2,3( )
)
通讯作者:
祝建波,吴燕民
作者简介:李相吴 E-mail:1003786451@qq.com;
基金资助:CLC Number:
Xiangwu LI, Ziyang LIU, Yujun XU, Jianbo ZHU, Yanmin WU. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis[J]. Journal of Agricultural Science and Technology, 2024, 26(1): 78-88.
李相吴, 刘自扬, 徐玉俊, 祝建波, 吴燕民. 真菌诱导子调控紫草素合成的分子机制探究[J]. 中国农业科技导报, 2024, 26(1): 78-88.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2022.0622
处理 Treatment | 样品编号 Sample ID | 总碱基数 Base number | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| F20 | F20-1 | 6 293 089 734 | 43.61 | 92.39 |
| F20-2 | 6 422 595 670 | 43.56 | 93.06 | |
| F20-3 | 7 834 038 640 | 43.62 | 92.46 | |
| R20 | R20-1 | 6 872 640 242 | 43.61 | 93.02 |
| R20-2 | 7 140 087 048 | 43.71 | 93.05 | |
| R20-3 | 6 946 752 472 | 43.66 | 92.70 | |
| CK | CK-1 | 7 792 349 390 | 43.66 | 92.82 |
| CK-2 | 6 870 409 240 | 43.68 | 92.81 | |
| CK-3 | 5 930 234 288 | 43.68 | 92.44 |
Table 1 sample sequencing data evaluation
处理 Treatment | 样品编号 Sample ID | 总碱基数 Base number | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| F20 | F20-1 | 6 293 089 734 | 43.61 | 92.39 |
| F20-2 | 6 422 595 670 | 43.56 | 93.06 | |
| F20-3 | 7 834 038 640 | 43.62 | 92.46 | |
| R20 | R20-1 | 6 872 640 242 | 43.61 | 93.02 |
| R20-2 | 7 140 087 048 | 43.71 | 93.05 | |
| R20-3 | 6 946 752 472 | 43.66 | 92.70 | |
| CK | CK-1 | 7 792 349 390 | 43.66 | 92.82 |
| CK-2 | 6 870 409 240 | 43.68 | 92.81 | |
| CK-3 | 5 930 234 288 | 43.68 | 92.44 |
处理 Treatment | 样品编号 Sample ID | Read数 Reads number | 匹配Read数 Mapped reads number | 匹配率 Mapped ratio/% |
|---|---|---|---|---|
| F20 | F20-1 | 21 096 427 | 15 558 872 | 73.75 |
| F20-2 | 21 504 765 | 16 195 509 | 75.31 | |
| F20-3 | 26 236 242 | 19 430 135 | 74.06 | |
| R20 | R20-1 | 23 005 518 | 17 277 331 | 75.10 |
| R20-2 | 23 913 025 | 17 644 691 | 73.79 | |
| R20-3 | 23 283 171 | 17 129 464 | 73.57 | |
| CK | CK-1 | 26 093 101 | 19 723 787 | 75.59 |
| CK-2 | 23 036 789 | 17 376 776 | 75.43 | |
| CK-3 | 19 879 659 | 14 903 996 | 74.97 |
Table 2 Comparison of sequencing data with assembly results
处理 Treatment | 样品编号 Sample ID | Read数 Reads number | 匹配Read数 Mapped reads number | 匹配率 Mapped ratio/% |
|---|---|---|---|---|
| F20 | F20-1 | 21 096 427 | 15 558 872 | 73.75 |
| F20-2 | 21 504 765 | 16 195 509 | 75.31 | |
| F20-3 | 26 236 242 | 19 430 135 | 74.06 | |
| R20 | R20-1 | 23 005 518 | 17 277 331 | 75.10 |
| R20-2 | 23 913 025 | 17 644 691 | 73.79 | |
| R20-3 | 23 283 171 | 17 129 464 | 73.57 | |
| CK | CK-1 | 26 093 101 | 19 723 787 | 75.59 |
| CK-2 | 23 036 789 | 17 376 776 | 75.43 | |
| CK-3 | 19 879 659 | 14 903 996 | 74.97 |
| 数据库名称Database name | 被注释到的差异基因数量 Annotated number of DGEs | |
|---|---|---|
| F20/CK | R20/CK | |
| COG | 517 | 317 |
| GO | 1 276 | 785 |
| KEGG | 1 063 | 651 |
| KOG | 758 | 439 |
| Pfam | 1 294 | 775 |
| Swiss-Prot | 1 188 | 731 |
| NR | 1 555 | 943 |
Table 3 Numbers of functional annotated DEGs
| 数据库名称Database name | 被注释到的差异基因数量 Annotated number of DGEs | |
|---|---|---|
| F20/CK | R20/CK | |
| COG | 517 | 317 |
| GO | 1 276 | 785 |
| KEGG | 1 063 | 651 |
| KOG | 758 | 439 |
| Pfam | 1 294 | 775 |
| Swiss-Prot | 1 188 | 731 |
| NR | 1 555 | 943 |
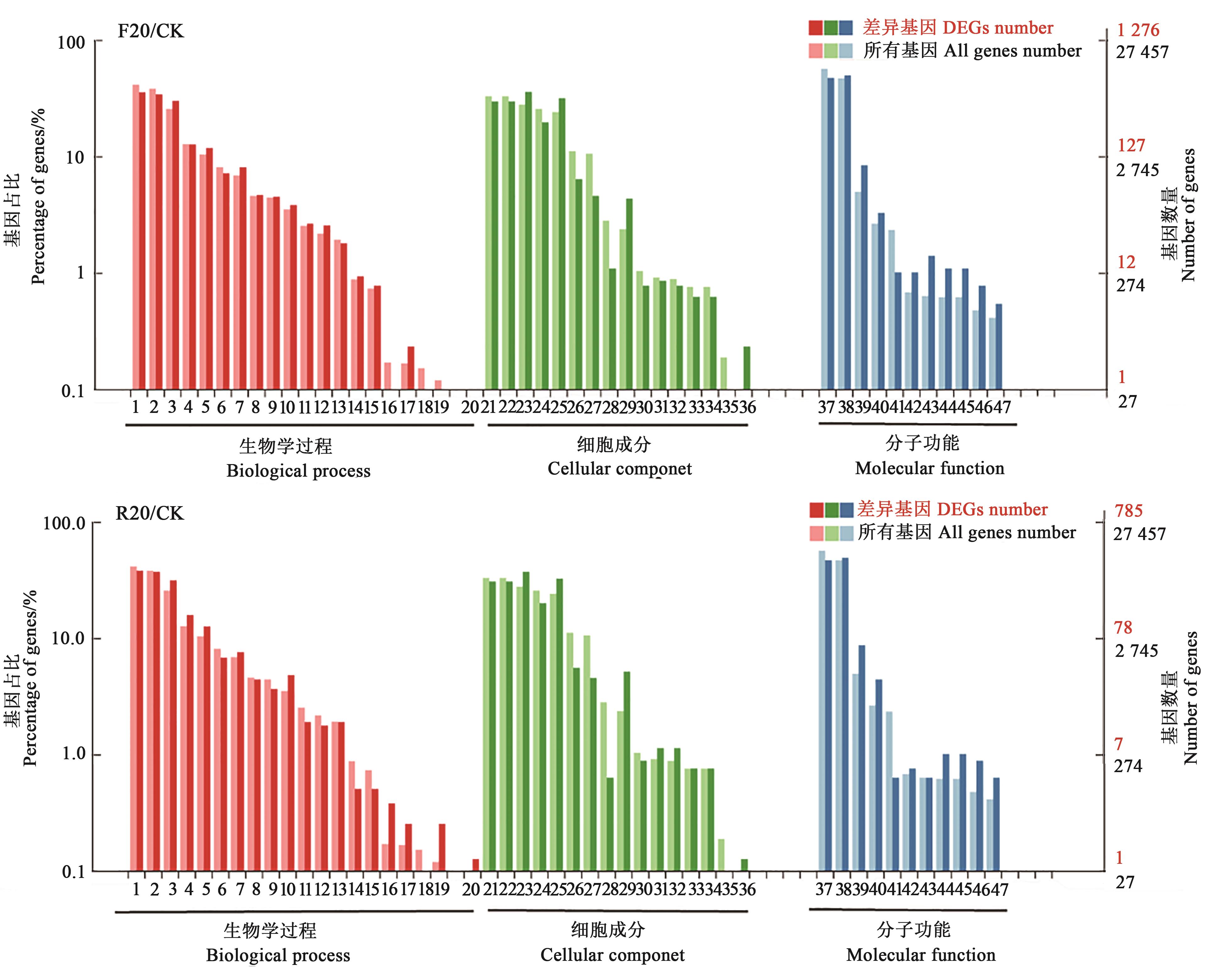
Fig. 2 GO functional annotation of differential expressed genesNote:1—Cellular process; 2—Metabolic process; 3—Single-organism process; 4—Biological regulation; 5—Response to stimulus; 6—Cellular component organization or biogenesis; 7—Localization; 8—Developmental process; 9—Multicellular organismal process; 10—Signaling; 11—Reproductive process; 12—Multi-organism process; 13—Reproduction; 14—Growth; 15—Immune system process; 16—Rhythmic process; 17—Locomotion; 18—Biological adhesion; 19—Detoxification; 20—Cell aggregation; 21—Cell; 22—Cell part; 23—membrane; 24—Organelle; 25—Membrane part; 26—Organelle part; 27—Macromolecular complex; 28—Membrane-enclosed lumen; 29—Extracellular region; 30—Extracellular region part; 31—Cell junction; 32—Symplast; 33—Other organism; 34—Other organism part; 35—Supramolecular complex; 36—Nucleoid; 37—Binding; 38—Catalytic activity; 39—Transporter activity; 40—Nucleic acid binding transcription factor activity; 41—Structural molecule activity; 42—Electron carrier activity; 43—Antioxidant activity; 44—Molecular transducer activity; 45—Signal transducer activity; 46—Transcription factor activity protein binding; 47—Molecular function regulator.
家族 Family | 基因ID Gene ID | log2FC | |
|---|---|---|---|
| F20/CK | R20/CK | ||
| TRAF | c65742.graph_c0 | 2.51 | 2.01 |
| Tify | c54302.graph_c0 | 1.59 | 0.96 |
| SRS | c54737.graph_c1 | -4.53 | -2.95 |
| MYB-related | c58018.graph_c0 | 4.26 | 2.80 |
| MYB | c62808.graph_c0 | 1.59 | 1.15 |
| LOB | c48779.graph_c0 | 1.71 | 1.28 |
| HB-other | c47461.graph_c0 | -2.71 | -1.85 |
| HB-BELL | c58247.graph_c0 | 1.70 | 1.54 |
| GARP-G2-like | c65836.graph_c0 | -1.95 | -1.23 |
| GARP-G2-like | c53563.graph_c1 | -1.78 | -1.82 |
| C2H2 | c56397.graph_c0 | -2.06 | -1.52 |
| C2C2-YABBY | c28651.graph_c0 | -3.50 | -2.26 |
| C2C2-Dof | c53061.graph_c0 | 1.71 | 1.50 |
| bHLH | c59392.graph_c0 | 2.16 | 1.43 |
| bHLH | c54786.graph_c0 | 1.65 | 1.66 |
| bHLH | c53656.graph_c0 | -6.60 | -3.19 |
| bHLH | c47522.graph_c0 | -2.99 | -1.71 |
| AP2/ERF-ERF | c60825.graph_c0 | -1.78 | -1.17 |
| AP2/ERF-ERF | c57468.graph_c0 | 2.29 | 1.93 |
| AP2/ERF-ERF | c55718.graph_c0 | -1.68 | -1.33 |
Table 4 Some differentially expressed transcription factor (top 20)
家族 Family | 基因ID Gene ID | log2FC | |
|---|---|---|---|
| F20/CK | R20/CK | ||
| TRAF | c65742.graph_c0 | 2.51 | 2.01 |
| Tify | c54302.graph_c0 | 1.59 | 0.96 |
| SRS | c54737.graph_c1 | -4.53 | -2.95 |
| MYB-related | c58018.graph_c0 | 4.26 | 2.80 |
| MYB | c62808.graph_c0 | 1.59 | 1.15 |
| LOB | c48779.graph_c0 | 1.71 | 1.28 |
| HB-other | c47461.graph_c0 | -2.71 | -1.85 |
| HB-BELL | c58247.graph_c0 | 1.70 | 1.54 |
| GARP-G2-like | c65836.graph_c0 | -1.95 | -1.23 |
| GARP-G2-like | c53563.graph_c1 | -1.78 | -1.82 |
| C2H2 | c56397.graph_c0 | -2.06 | -1.52 |
| C2C2-YABBY | c28651.graph_c0 | -3.50 | -2.26 |
| C2C2-Dof | c53061.graph_c0 | 1.71 | 1.50 |
| bHLH | c59392.graph_c0 | 2.16 | 1.43 |
| bHLH | c54786.graph_c0 | 1.65 | 1.66 |
| bHLH | c53656.graph_c0 | -6.60 | -3.19 |
| bHLH | c47522.graph_c0 | -2.99 | -1.71 |
| AP2/ERF-ERF | c60825.graph_c0 | -1.78 | -1.17 |
| AP2/ERF-ERF | c57468.graph_c0 | 2.29 | 1.93 |
| AP2/ERF-ERF | c55718.graph_c0 | -1.68 | -1.33 |
| 1 | 国家药典委员会.中华人民共和国药典-一部[M].北京:中国医药科技出版社,2020:355-356. |
| National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China-part1 [M]. Beijing: Chinese Medicine Science and Technology Press, 2020:355-356. | |
| 2 | WANG Y, ZHU Y, XIAO L, et al... Meroterpenoids isolated from Arnebia euchroma (Royle) Johnst. and their cytotoxic activity in human hepatocellular carcinoma cells [J]. Fitoterapia, 2018, 131(11):236-244. |
| 3 | CRUICKSHANK I, PERRIN D R. The isolation and partial characterization of monilicolin A, a polypeptide with phaseollin-inducing activity from Monilinia fructicola [J]. Life Sci., 1968, 7(10):449-458. |
| 4 | OKSMAN-CALDENTEY K, VERPOORTE R, VAN DER HEIJDEN R, et al... Engineering the plant cell factory for secondary metabolite production [J]. Transgenic Res., 2000, 9(4):323-343. |
| 5 | WANG Y, DAI C C, CAO J L, et al.. Comparison of the effects of fungal endophyte Gilmaniella sp. and its elicitor on atractylodes lancea plantlets [J]. World J. Microbiol. Biotechnol., 2011, 28(2):575-584. |
| 6 | ARGHAVANI P, HAGHBEEN K, MOUSAVI A. Enhancement of shikalkin production in Arnebia euchroma callus by a fungal elicitor, Rhizoctonia solani [J]. Iranian J. Biotechnol., 2015, 13(4):10-16. |
| 7 | 晏琼,胡宗定,吴建勇.生物与非生物诱导子协同作用对丹参毛状根培养生产丹参酮的影响[J].中国中药杂志,2006(3):188-191. |
| YAN Q, HU Z D, WU J Y. Synergistic effects of biotic and abiotic elicitors on the production of tanshinones in Salvia miltiorrhiza hairy root culture [J]. China J. Chin. Materia Medica, 2006(3):188-191. | |
| 8 | 田佩雯.白及内生真菌诱导子对宿主生长和主要活性物质的影响及调控[D].南宁:广西大学,2019. |
| TIAN P W. Effects and regulation of endophytic fungal elicitors from Bletilla striata on host growthand main substances [D]. Nanning: Guangxi University, 2019. | |
| 9 | 饶龙兵,杨汉波,郭洪英,等.不同倍性桤木属植物的转录组测序和分析[J].分子植物育种.2016,14(11):3047-3055. |
| RAO L B, YANG H B, GUO H Y, et al.. Analysis on transcriptome sequenced for alnus plants with different ploidy [J]. Mol. Plant Breed., 2016, 14(11):3047-3055. | |
| 10 | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 [J/OL]. Genome Biol., 2014, 15(12):550 [2022-06-02]. |
| 11 | GAISSER S, HEIDE L. Inhibition and regulation of shikonin biosynthesis in suspension cultures of Lithospermum [J]. Phytochemistry, 1996, 41(4):1065-1072. |
| 12 | SYKŁOWSKA-BARANEK K, PIETROSIUK A, NALIWAJSKI M R, et al.. Effect of L-phenylalanine on PAL activity and production of naphthoquinone pigments in suspension cultures of Arnebia euchroma (Royle) Johnst [J]. In Vitro Cellular Dev. Biol. Plant, 2012, 48(5):555-564. |
| 13 | YAZAKI K, KUNIHISA M, FUJISAKI T, et al.. Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon: cloning and characterization of a key enzyme in shikonin biosynthesis [J]. J. Biol. Chem., 2002, 277(8):6240-6246. |
| 14 | WANG S, WANG R S, LIU T G, et al.. CYP76B74 catalyzes the 3’’-hydroxylation of geranylhydroquinone in shikonin biosynthesis [J]. Plant Physiol. (Bethesda), 2019, 179(2):402-414. |
| 15 | 梁玖雯,李锬,王瑞杉,等.新疆紫草中2条CYP450基因的干扰毛状根体系的建立及其影响研究[J].中国中药杂志,2020,45(14):3422-3431. |
| LIANG J W, LI T, WANG R S, et al.. Establishment of RNA interfered hairy root system of two CYP450 genes in Arnebia euchroma and its influence [J]. China J. Chin. Materia Medica, 2020, 45(14):3422-3431. | |
| 16 | LIAO M, ZENG C, LIANG F. Two new dimeric naphthoquinones from Arnebia euchroma [J]. Phytochem. Letters, 2020, 37(1):106-109. |
| 17 | YAZAKI K, MATSUOKA H, SHIMOMURA K, et al.. A novel dark-inducible protein, LeDI-2, and its involvement in root-specific secondary metabolism in Lithospermum erythrorhizon [J]. Plant Physiol. (Bethesda), 2001, 125(3):1831-1841. |
| 18 | YAMAMURA Y N C U, SAHIN F P, NAGATSU A, et al.. Molecular cloning and characterization of a cDNA encoding a novel apoplastic protein preferentially expressed in a shikonin-producing callus strain of Lithospermum erythrorhizon [J]. Plant Cell Physiol., 2003, 44(4):437-446. |
| 19 | SAHA S, PAL D. Elicitor Signal Transduction Leading to the Production of Plant Secondary Metabolites [M]. Cham: Springer International Publishing, 2020:1-39. |
| 20 | 周雅涵.水杨酸、膜醭毕赤酵母、壳寡糖诱导柑橘果实抗病性及其生物学机制研究[D].重庆:西南大学,2017. |
| ZHOU Y H. Salicylic acid, Pichia membranaefaciens and oligochitosan induced disease resistance of citrus fruit and the possible biological mechanisms involved [D]. Chongqing: Southwest University, 2017. | |
| 21 | 瞿巾卓.酿酒葡萄细胞对内生真菌诱导子的代谢响应与机制[D].昆明:云南大学,2020. |
| JU J Z. Metabolic response and mechanism of Wine grape cells to elicitors from fungal endophytes [D]. Kunming: Yunnan University, 2020 | |
| 22 | 张明菊,朱莉,夏启中.植物激素对胁迫反应调控的研究进展[J].湖北大学学报(自然科学版).2021,43(3):242-253. |
| ZHANG M J, ZHU L, XIA Q ZAND. Research progress on the regulation of plant hormones to stress responses [J]. J. Hubei Univ., 2021, 43(3):242-253. | |
| 23 | SHAH L, ALI A, ZHU Y L, et al.. Wheat defense response to Fusarium head blight and possibilities of its improvement [J]. Physiol. Mol. Plant Pathol., 2017, 98(2):9-17. |
| 24 | NEMESIO-GORRIZ M, BLAIR P B, DALMAN K, et al.. Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway [J/OL]. Front. Plant Sci., 2017, 8:305 [2022-06-02]. . |
| 25 | GRAEFF M, STRAUB D, EGUEN T, et al.. Microprotein-mediated recruitment of constans into a topless trimeric complex represses flowering in Arabidopsis [J/OL]. PLoS Genet., 2016, 12(3):e1005959 [2022-06-02]. . |
| 26 | DENG B, HUANG Z, GE F, et al.. An AP2/ERF family transcription factor PnERF1 raised the biosynthesis of saponins in panax notoginseng [J]. J. Plant Growth Regul., 2017, 36(3):691-701. |
| 27 | GAUTAM J K, GIRI M K, SINGH D, et al.. MYC2 influences salicylic acid biosynthesis and defense against bacterial pathogens in Arabidopsis thaliana [J]. Physiol. Plantarum., 2021, 173(4):2248-2261. |
| 28 | KAZAN K, MANNERS J M. MYC2: the master in action [J]. Mol. Plant, 2013, 6(3):686-703. |
| 29 | WANG F, ZHU H, CHEN D, et al.. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana [J]. Plant Cell. Tissue Organ Cult., 2016, 125(2):387-398. |
| 30 | ZHANG M, LI S T, NIE L, et al.. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis [J]. Plant Mol. Biol., 2015, 89(4-5):463-473. |
| 31 | EL-SAYED A S A, MOHAMED N Z, SAFAN S, et al.. Restoring the taxol biosynthetic machinery of aspergillus terreus by Podocarpus gracilior pilger microbiome, with retrieving the ribosome biogenesis proteins of WD40 superfamily [J]. Sci. Rep., 2019, 9(1):11512-11534. |
| 32 | HAN Z, YANG T, GUO Y, et al.. The transcription factor PagLBD3 contributes to the regulation of secondary growth in Populus [J]. J. Exp. Bot., 2021, 72(20):7092-7106. |
| 33 | TANG X M, WANG X, HUANG Y, et al... Natural variations of TFIIAγ gene and LOB1 promoter contribute to citrus canker disease resistance in Atalantia buxifolia [J]. PLoS Genetics. 2021, 17(1):e1009316 [2022-06-02]. . |
| 34 | HAO H, LEI C, DONG Q, et al... Effects of exogenous methyl jasmonate on the biosynthesis of shikonin derivatives in callus tissues of Arnebia euchroma [J]. Appl. Biochem. Biotechnol., 2014, 173(8):2198-2210. |
| 35 | WANG C G, WU J Y, MEI X G. Enhancement of taxol production and excretion in Taxus chinensis cell culture by fungal elicitation and medium renewal [J]. Appl. Microbiol. Biotechnol., 2001, 55(4):404-410. |
| [1] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [2] | Xinxin WANG, Ziwei WU, Ting FANG, Qiao MOU, Fang WANG, Han ZHAO, Zhiqiang DU, Caixia YANG. Vitamin C Modulates Function and Gene Expression of Mouse TM4 Sertoli Cells [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 81-89. |
| [3] | Qian ZHANG, Lina MEN, Yiran LI, Qiao LIU, Angie DENG, Xiaowen HU, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Differential Expression Paradigm of Chemoreceptor Genes Between Males and Females at Different Developmental Stages of Carposina sasakii Matsumura [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 151-162. |
| [4] | Yiwei LU, Xueyan XIA, Yu ZHAO, Jihan CUI, Meng LIU, Meihong HUANG, Cheng CHU, Jianjun LIU, Shunguo LI. Transcriptome Profiling and Gene Mining of Millet Response to Potassium Deficiency Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 30-44. |
| [5] | Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43. |
| [6] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [7] | Zhan ZHAO, Xiaoting WANG, Lifeng ZHANG, Jinhe ZHAO, Yuhong YU, Junhua LI, Zhanqing WU. Transcriptome Analysis of Watermelon Responses to Low Nitrogen Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 30-38. |
| [8] | Wei JI, Ying FAN, Jiaxing HUANG, Huipeng YANG, Jin XU, Xiaoying LI, Yueqin GUO, Yueguo WU, Jilian LI, Jun YAO. Transcriptome Analysis of Stigma Response Mechanism of Blueberry with Different Pollination Intensity [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 71-82. |
| [9] | Wanjing XU, Fang PENG, Doudou ZHAO, Jiaojiao LUO, Shan TAO, Hailang LIAO, Changqing MAO, Yu WU, Xiu ZHU, Zhengjun XU, Chao ZHANG. Analysis of Response Mechanism of Ligusticum chuanxiong to Cadmium Stress Based on Transcriptome and Metabolome [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 98-109. |
| [10] | Xiaoran WANG, Xiaoyu LI, Hui SUN, Haidong YU, Yongchun SHI. Transcriptome Analysis of Tobacco Leaves Under Boron Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 53-64. |
| [11] | Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86. |
| [12] | Lan MA, Qing PENG, Xiaoqing XU, Shuo YANG, Yuwei ZHANG, Dandan TIAN, Linbo SHI, Bo SHI, Yu QIAO. Gene Expression in Escherichia coli O157∶H7 Biofilms [J]. Journal of Agricultural Science and Technology, 2023, 25(6): 71-88. |
| [13] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [14] | Ruifeng GUO, Yuemei REN, Zhong YANG, Guishan LIU, Guangbing REN, Shou ZHANG, Wenjuan ZHU. Transcriptomic Analysis of Mechanism of Foxtail Millet Male Infertility Induced by Glyphosate Ammonium Salt [J]. Journal of Agricultural Science and Technology, 2022, 24(10): 35-43. |
| [15] | LI Shuxin, ZHANG Hao, ZHENG Housheng, ZHENG Peihe, PANG Shifeng, XU Shiquan. Transcriptome Analysis of Phenotypic Differences Between Ermaya and Changbo of Forest Cultivated Panax ginseng [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 56-68. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号