




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (7): 77-86.DOI: 10.13304/j.nykjdb.2022.0170
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Yunsheng WANG( ), Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG(
), Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG( )
)
Received:2022-03-07
Accepted:2022-04-20
Online:2023-07-15
Published:2023-08-25
Contact:
Chuanbo ZHANG
通讯作者:
张传博
作者简介:王云胜 E-mail:1710818668@qq.com;
基金资助:CLC Number:
Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus[J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86.
王云胜, 陈银翠, 程在, 张锦, 张传博. 过表达veA基因对冠突散囊菌次级代谢的影响[J]. 中国农业科技导报, 2023, 25(7): 77-86.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2022.0170
引物 Primer | 序列 Sequence (5’-3’) |
|---|---|
| veAF | AGACATCACCATGGTAGATCTATGGCGACGCGAATGCCC |
| veAR | GGGGAAATTCGAGCTGGTCACCTTAACTAGAAAAAGCCGGCGG |
| GAPDHF | CTGCCGTATCGAGAAGGGTG |
| GAPDHR | GATGAAGTTGGGGTTGAGGG |
| QveAF | CGACATCACCTTTGCCTAC |
| QveAR | TCGGGAAAGATGAAGTAGC |
Table 1 Primers used in this study
引物 Primer | 序列 Sequence (5’-3’) |
|---|---|
| veAF | AGACATCACCATGGTAGATCTATGGCGACGCGAATGCCC |
| veAR | GGGGAAATTCGAGCTGGTCACCTTAACTAGAAAAAGCCGGCGG |
| GAPDHF | CTGCCGTATCGAGAAGGGTG |
| GAPDHR | GATGAAGTTGGGGTTGAGGG |
| QveAF | CGACATCACCTTTGCCTAC |
| QveAR | TCGGGAAAGATGAAGTAGC |
样品 Sample | 原始数据 Raw reads | 质控数据 Clean reads | 错配率 Error rate/% | Q20/% | Q30/% |
|---|---|---|---|---|---|
| OE::veA1_1 | 53 215 236 | 52 880 844 | 0.023 9 | 98.44 | 95.28 |
| OE::veA1_2 | 58 759 294 | 58 355 148 | 0.024 0 | 98.4 | 95.21 |
| OE::veA1_3 | 56 979 242 | 56 535 564 | 0.024 1 | 98.36 | 95.11 |
| WT1 | 52 014 002 | 51 659 080 | 0.024 0 | 98.41 | 95.24 |
| WT2 | 57 594 222 | 57 246 210 | 0.023 9 | 98.43 | 95.26 |
| WT3 | 58 672 768 | 58 282 756 | 0.023 9 | 98.42 | 95.25 |
Table 2 Transcriptome sequencing data statistics
样品 Sample | 原始数据 Raw reads | 质控数据 Clean reads | 错配率 Error rate/% | Q20/% | Q30/% |
|---|---|---|---|---|---|
| OE::veA1_1 | 53 215 236 | 52 880 844 | 0.023 9 | 98.44 | 95.28 |
| OE::veA1_2 | 58 759 294 | 58 355 148 | 0.024 0 | 98.4 | 95.21 |
| OE::veA1_3 | 56 979 242 | 56 535 564 | 0.024 1 | 98.36 | 95.11 |
| WT1 | 52 014 002 | 51 659 080 | 0.024 0 | 98.41 | 95.24 |
| WT2 | 57 594 222 | 57 246 210 | 0.023 9 | 98.43 | 95.26 |
| WT3 | 58 672 768 | 58 282 756 | 0.023 9 | 98.42 | 95.25 |
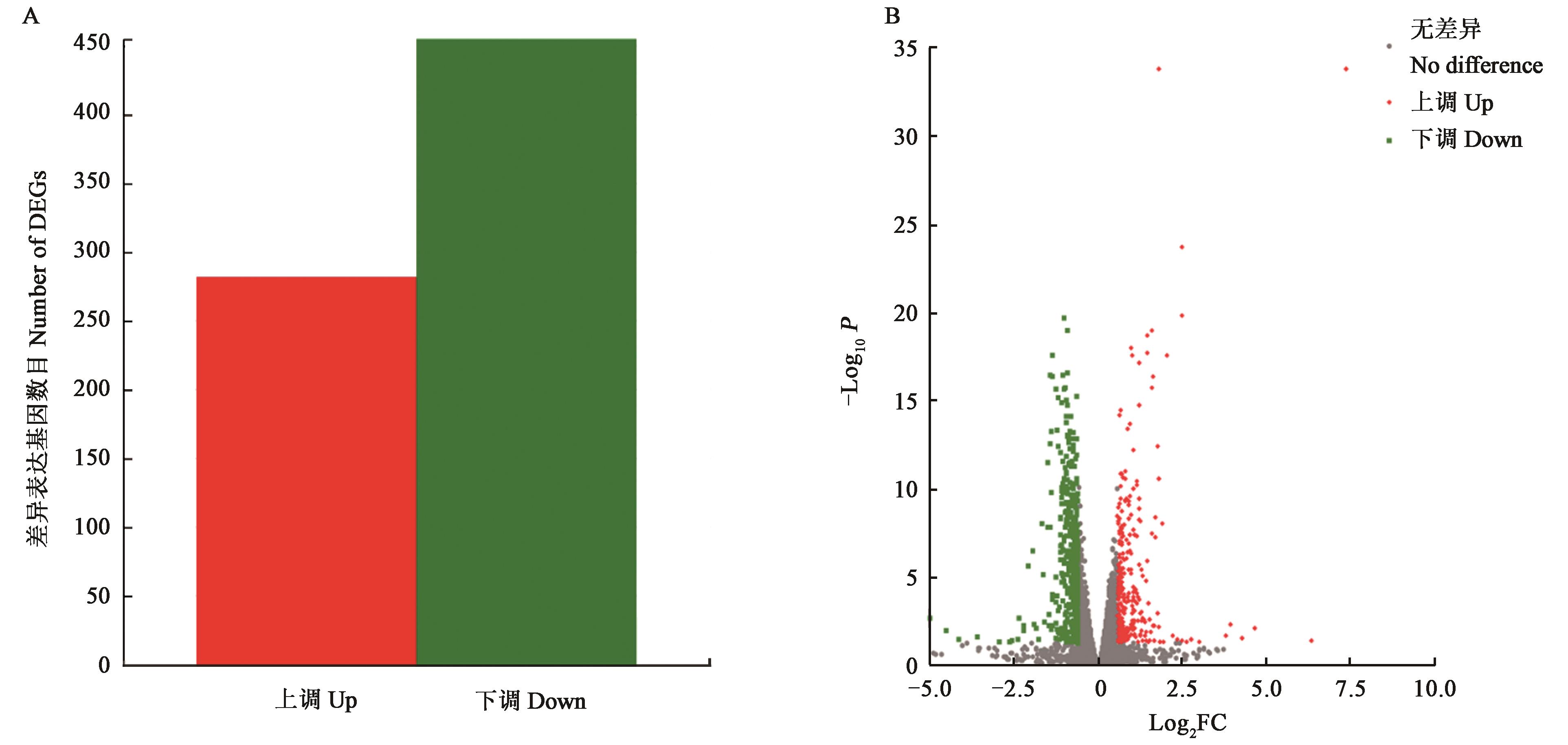
Fig. 3 Differentially expressed genes between WT and OE::veA1A: Histogram of differentially expressed genes; B: Volcano map of differentially expressed genes
基因编号 Gene ID | 功能注释 Function annotation | 变化倍数 Fold change | 调控 Regulate |
|---|---|---|---|
| SI65_05759 | 氧化还原酶 Flavoprotein-ubiquinone oxidoreductase | 1.01ns | 上调Up |
| SI65_05760 | 反转录酶 Reverse transcriptase | 0.61ns | 下调Down |
| SI65_05761 | TATA框结合蛋白 TATA-box-binding protein | 0.72ns | 下调Down |
| SI65_05762 | L-甲基哌啶氧化酶 L-pipecolate oxidase | 1.50* | 上调Up |
| SI65_05763 | 甲基转移酶 Methyltransferase | 1.05ns | 上调Up |
| SI65_05764 | 假设蛋白 Hypothetical protein | 1.45ns | 上调Up |
| SI65_05765 | C6指域转录因子 C6 finger domain transcription factor | 1.25ns | 上调Up |
| SI65_05766 | NADH-生化色素b5还原酶 NADH-cytochrome b5 reductase | 1.27ns | 上调 Up |
| SI65_05767 | 甲基转移酶 Methyltransferase | 1.72* | 上调Up |
| SI65_05768 | 羧基酸合成酶 Carboxylic acid synthase | 2.02* | 上调Up |
| SI65_05769 | 金属化合物-β-内酰胺酶 Metallo-beta-lactamase | 2.31* | 上调Up |
| SI65_05770 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 1.92ns | 上调Up |
| SI65_05771 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 2.47* | 上调 Up |
| SI65_05772 | 氧化还原酶 Oxidoreductase | 2.67* | Up 上调 |
| SI65_05773 | 乙酰基转移酶 Acetyltransferase | 2.74* | Up 上调 |
| SI65_05774 | 假定蛋白 Hypothetical protein | NA | NA |
| SI65_05775 | 假定蛋白 Hypothetical protein | 0.55* | Down 下调 |
| SI65_05776 | 转录因子 Transcription factor | 0.64* | Down 下调 |
Table 3 Gene expression and functional annotation in secondary metabolite gene cluster 32
基因编号 Gene ID | 功能注释 Function annotation | 变化倍数 Fold change | 调控 Regulate |
|---|---|---|---|
| SI65_05759 | 氧化还原酶 Flavoprotein-ubiquinone oxidoreductase | 1.01ns | 上调Up |
| SI65_05760 | 反转录酶 Reverse transcriptase | 0.61ns | 下调Down |
| SI65_05761 | TATA框结合蛋白 TATA-box-binding protein | 0.72ns | 下调Down |
| SI65_05762 | L-甲基哌啶氧化酶 L-pipecolate oxidase | 1.50* | 上调Up |
| SI65_05763 | 甲基转移酶 Methyltransferase | 1.05ns | 上调Up |
| SI65_05764 | 假设蛋白 Hypothetical protein | 1.45ns | 上调Up |
| SI65_05765 | C6指域转录因子 C6 finger domain transcription factor | 1.25ns | 上调Up |
| SI65_05766 | NADH-生化色素b5还原酶 NADH-cytochrome b5 reductase | 1.27ns | 上调 Up |
| SI65_05767 | 甲基转移酶 Methyltransferase | 1.72* | 上调Up |
| SI65_05768 | 羧基酸合成酶 Carboxylic acid synthase | 2.02* | 上调Up |
| SI65_05769 | 金属化合物-β-内酰胺酶 Metallo-beta-lactamase | 2.31* | 上调Up |
| SI65_05770 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 1.92ns | 上调Up |
| SI65_05771 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 2.47* | 上调 Up |
| SI65_05772 | 氧化还原酶 Oxidoreductase | 2.67* | Up 上调 |
| SI65_05773 | 乙酰基转移酶 Acetyltransferase | 2.74* | Up 上调 |
| SI65_05774 | 假定蛋白 Hypothetical protein | NA | NA |
| SI65_05775 | 假定蛋白 Hypothetical protein | 0.55* | Down 下调 |
| SI65_05776 | 转录因子 Transcription factor | 0.64* | Down 下调 |
| 1 | BOK J, HOFFMEISTER D, MAGGIO-HALL L, et al.. Genomic mining for Aspergillus natural products [J]. Chem. Biol., 2006, 13(1):7-31. |
| 2 | RAJA H A, MILLER A N, PERACE C J, et al.. Fungal identification using molecular tools: a primer for the natural products research community [J]. J. Nat. Prod., 2017, 80(3):756-770. |
| 3 | ZHANG X J, ZHU Y Y, BAO L F, et al.. Putative methyltransferase laea and transcription factor CREA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression [J]. Fungal. Genet. Biol., 2016, 94:32-46. |
| 4 | BAYRAM O, BRAUS G H. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins [J]. FEMS Microbiol. Lett., 2012, 36(1):1-24. |
| 5 | KELLER N. Fungal secondary metabolism: regulation, function and drug discovery [J]. Nat. Rev. Microbiol., 2019, 17(3):167-180. |
| 6 | CHIANG Y M, OAKLEY C E, AHUJA M E, et al.. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus Nidulans [J]. J. Am. Chem. Soc., 2013, 135(20):7720-7731. |
| 7 | YAEGASHI J, OAKLEY B, WANG C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans [J]. J. Appl. Microbiol., 2014, 41(2):433-442. |
| 8 | SARIKAYA-BATRAM Ö, PALMER JM, KELLER N, et al.. One juliet and four romeos: vea and its methyltransferases [J/OL]. Front. Microbiol., 2015, 6:1 [2022-02-10]. . |
| 9 | KATO N, BROOKS W, CALVO A. The expression of sterigmatocystin and penicillin genes in Aspergillus Nidulans is controlled by vea, a gene required for sexual development [J]. Eukaryot. Cell, 2003, 2(6):1178-1186. |
| 10 | MARUI J, OHASHI-KUNIHIRO S, ANDO T, et al.. Penicillin biosynthesis in Aspergillus Oryzae and its overproduction by genetic engineering [J]. J. Biosci. Bioeng., 2010, 110(1):8-11. |
| 11 | 曹双,郑跃亮,张虎,等.过表达veA对橘青霉美伐他汀生物合成的影响[J].西南师范大学学报(自然科学版),2016,41(6):67-73. |
| CAO S, ZHENG Y L, ZHANG H, et al.. On investigation of veA gene in regulating mevastatin biosynthesis, conidia development in Penicillium citrinum [J]. J. Southwest China Normal Univ. (Nat. Sci.), 2016, 41(6):67-73. | |
| 12 | 张月,崔旋旋,刘英学,等.茯砖茶中冠突散囊菌的分离鉴定及其发酵工艺和生物活性研究[J].食品与发酵工业,2020,418(22):202-207. |
| ZHANG Y, CUI X X, LIU Y X, et al.. Isolation, identification, fermentation technology and bioactivity of Eurotium cristatum in Fuzhuan brick tea [J]. Food. Ferment. Ind., 2020, 46(22):202-207. | |
| 13 | AN T T, CHEN M X, ZU Z Q, et al.. Untargeted and targeted metabolomics reveal changes in the chemical constituents of instant dark tea during liquid-state fermentation by Eurotium cristatum [J]. Food. Res. Int., 2021, 148:110-125. |
| 14 | 陈敏,谢发,游玲,等.冠突散囊菌发酵苦丁茶工艺研究[J].食品与发酵工业, 2020,402(6):224-228. |
| CHNEG M, XIE F, YOU L, et al.. Fermentation of ligustrum robustum by Eurotium cristatum [J]. Food. Ferment. Ind., 2020, 46(6):224-228. | |
| 15 | GE Y Y, WANG Y C, LIU Y X, et al.. Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus Aspergillus cristatus [J/OL]. BMC Genomics, 2016, 17(1): 428 [2022-02-10]. . |
| 16 | 周国庆.冠突曲霉LaeA基因功能的研究[D].贵阳:贵州大学,2018. |
| ZHOU G Q. Study on the function of LaeA gene in Aspergillus cristatum [D]. Guiyang: Guizhou University, 2018. | |
| 17 | TAN Y M, WANG H, WANG Y P, et al.. The role of the vea gene in adjusting developmental balance and environmental stress response in Aspergillus cristatus [J]. Fungal. Biol., 2018, 122(10):952-964. |
| 18 | WANG Y P, TAN Y M, WANG Y C, et al.. Role of Acndta in cleistothecium formation, osmotic stress response, pigmentation and carbon metabolism of Aspergillus cristatus [J]. Fungal. Biol., 2021, 125(10):749-763. |
| 19 | 刘逸梅.冠突曲霉钙信号调控基因Fig的功能研究[D].贵阳:贵州大学, 2019. |
| LIU YM. Study on the function of Fig gene regulating calcium signal in Aspergillus cristatum [D]. Guiyang: Guizhou University, 2019. | |
| 20 | 朱思远,徐岩,喻晓蔚.农杆菌介导转化黑曲霉条件优化及脂肪酶表达[J].食品与生物技术学报,2020,39(5):51-58. |
| ZHU S Y, XU Y, YU X Y. Optimization of agrobacterium tumefaciens-mediated transformation of Aspergillus niger and expression of lipase [J]. J. Food Sci. Biotechnol., 2020, 39(5):51-58. | |
| 21 | 邵蕾,谭玉梅,任春光,等.转录因子ACZ影响冠突曲霉色素及产孢数量的研究[J].基因组学与应用生物学,2019,38(5):2041-2048. |
| SHAO L, TAN Y M, REN C G, et al.. Study on the transcription factor ACZ affecting the number of spores and pigmentation of Asperillus cristatus [J]. Genom. Appl. Biol., 2019, 38(5):2041-2048. | |
| 22 | 向婷,于凤明,杨正军,等.冠突曲霉preA基因的功能分析[J].基因组学与应用生物学,2020,39(7):3070-3078. |
| XIANG T, YU F M, YANG Z J, et al.. Functional analysis on the preA gene in Aspergillus cristatus [J]. Genom. Appl. Biol., 2020, 39(7):3070-3078. | |
| 23 | 杜静.冠突散囊菌对中药材三七成分的转化及机理研究[D].贵阳:贵州师范大学,2020. |
| DU J. Biotransformation of bioactive compounds in Pannax notoginseng fermented by Eurotium cristatum and its mechanism [D]. Guiyang: Guizhou Normal University, 2020. | |
| 24 | 王琪琪.黑茶中散囊属真菌及其对茶叶品质提升研究[D].贵阳:贵州师范大学,2021. |
| WANG Q Q. The diversity of Eurotium sp . in dark tea and its effect on the quality improvement [D]. Guiyang: Guizhou Normal University, 2021. | |
| 25 | REN X X, WANG Y C, LIU Y X, et al.. Comparative transcriptome analysis of the calcium signaling and expression analysis of sodium/calcium exchanger in Aspergillus cristatus [J]. J. Basic. Microbiol., 2018, 58(1):76-87. |
| 26 | WANG D Y, TONG S M, GUAN Y Y, et al.. The velvet protein vea functions in asexual cycle, stress tolerance and transcriptional regulation of Beauveria bassiana [J]. Fungal. Genet. Biol., 2019, 127:1-11. |
| 27 | LIU H, SANG S L, WANG H, et al.. Vea comparative proteomic analysis reveals the regulatory network of the gene during asexual and sexual spore development of Aspergillus cristatus [J/OL]. Biosci. Rep., 2018, 38(4):67 [2022-02-10]. . |
| 28 | 张玉婷,李伟国,梁冬梅,等. P450s在萜类合成方面的合成生物学研究进展[J].中国生物工程杂志,2020,40(8):84-96. |
| ZHANG Y T, LI W G, LIANG D M, et al.. Research progress in synthetic biology of P450s in terpenoid synthesis [J]. Chin. J. Biotechnol., 2020, 40(8):84-96. | |
| 29 | CHANG M, KEASLING J. Production of isoprenoid pharmaceuticals by engineered microbes [J]. Nat. Chem. Biol., 2006, 2(12):674-681. |
| 30 | CARY J W, HAN Z, YIN Y, et al.. Transcriptome analysis of Aspergillus flavus reveals vea-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster [J]. Eukaryot. Cell., 2015, 14(10):983-997. |
| 31 | 张梦薇.过表达laeA和veA基因对黑曲霉生长和代谢的影响[D].天津:天津科技大学,2020. |
| ZHANG M W. Effects of overexpression of laeA and veA genes on growth and metabolism of Aspergillus niger [D]. Tianjin: Tianjin University of Science and Technology, 2020. | |
| 32 | KONG W P, HUANG C S, SHI J, et al.. Recycling of Chinese herb residues by endophytic and probiotic fungus Aspergillus cristatus Cb10002 for the production of medicinal valuable anthraquinones [J/OL]. Microb. Cell. Fact., 2019, 18(1):9 [2022-02-10]. . |
| [1] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [2] | Xinxin WANG, Ziwei WU, Ting FANG, Qiao MOU, Fang WANG, Han ZHAO, Zhiqiang DU, Caixia YANG. Vitamin C Modulates Function and Gene Expression of Mouse TM4 Sertoli Cells [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 81-89. |
| [3] | Qian ZHANG, Lina MEN, Yiran LI, Qiao LIU, Angie DENG, Xiaowen HU, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Differential Expression Paradigm of Chemoreceptor Genes Between Males and Females at Different Developmental Stages of Carposina sasakii Matsumura [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 151-162. |
| [4] | Yiwei LU, Xueyan XIA, Yu ZHAO, Jihan CUI, Meng LIU, Meihong HUANG, Cheng CHU, Jianjun LIU, Shunguo LI. Transcriptome Profiling and Gene Mining of Millet Response to Potassium Deficiency Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 30-44. |
| [5] | Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43. |
| [6] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [7] | Zhan ZHAO, Xiaoting WANG, Lifeng ZHANG, Jinhe ZHAO, Yuhong YU, Junhua LI, Zhanqing WU. Transcriptome Analysis of Watermelon Responses to Low Nitrogen Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 30-38. |
| [8] | Wei JI, Ying FAN, Jiaxing HUANG, Huipeng YANG, Jin XU, Xiaoying LI, Yueqin GUO, Yueguo WU, Jilian LI, Jun YAO. Transcriptome Analysis of Stigma Response Mechanism of Blueberry with Different Pollination Intensity [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 71-82. |
| [9] | Wanjing XU, Fang PENG, Doudou ZHAO, Jiaojiao LUO, Shan TAO, Hailang LIAO, Changqing MAO, Yu WU, Xiu ZHU, Zhengjun XU, Chao ZHANG. Analysis of Response Mechanism of Ligusticum chuanxiong to Cadmium Stress Based on Transcriptome and Metabolome [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 98-109. |
| [10] | Xiangwu LI, Ziyang LIU, Yujun XU, Jianbo ZHU, Yanmin WU. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 78-88. |
| [11] | Xiaoran WANG, Xiaoyu LI, Hui SUN, Haidong YU, Yongchun SHI. Transcriptome Analysis of Tobacco Leaves Under Boron Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 53-64. |
| [12] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [13] | LI Shuxin, ZHANG Hao, ZHENG Housheng, ZHENG Peihe, PANG Shifeng, XU Shiquan. Transcriptome Analysis of Phenotypic Differences Between Ermaya and Changbo of Forest Cultivated Panax ginseng [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 56-68. |
| [14] | LIU Yuan, ZHANG Xiuyan, XU Miaoyun, ZHENG Hongyan, ZOU Junjie, ZHANG Lan, WANG Lei. Global Small RNA Transcriptome Profiling of Rice Under Drought Stress [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 23-32. |
| [15] | ZHANG Wenyun1, ZHANG Jiancheng2, YAO Jingzhen2*. Comparative Transcriptome Analysis of Wheat Leaf in Response to Low Nitrogen Stress#br# [J]. Journal of Agricultural Science and Technology, 2020, 22(11): 26-34. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号