




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (2): 227-233.DOI: 10.13304/j.nykjdb.2021.0785
• 方法与技术创新 • 上一篇
胡秀文1( ), 邓波2(
), 邓波2( ), 王金斌3, 刘华3(
), 王金斌3, 刘华3( ), 唐雪明4(
), 唐雪明4( ), 王宇5, 曾海娟3, 蒋玮3, 李红6
), 王宇5, 曾海娟3, 蒋玮3, 李红6
收稿日期:2021-09-07
接受日期:2021-10-20
出版日期:2023-02-15
发布日期:2023-05-17
通讯作者:
刘华,唐雪明
作者简介:胡秀文 E-mail:849727644@qq.com基金资助:
Xiuwen HU1( ), Bo DENG2(
), Bo DENG2( ), Jinbin WANG3, Hua LIU3(
), Jinbin WANG3, Hua LIU3( ), Xueming TANG4(
), Xueming TANG4( ), Yu WANG5, Haijuan ZENG3, Wei JIANG3, Hong LI6
), Yu WANG5, Haijuan ZENG3, Wei JIANG3, Hong LI6
Received:2021-09-07
Accepted:2021-10-20
Online:2023-02-15
Published:2023-05-17
Contact:
Hua LIU,Xueming TANG
摘要:
核酸检测在农产品安全检测中的应用十分广泛,随着转基因作物的商业化种植,迫切需要一种快速、特异、灵敏的转基因作物检测方法。利用重组酶聚合酶扩增(recombinase polymerase amplification,RPA)技术对含有转基因产品及其制品中CP4-EPSPS基因进行检测,共设计5对引物来筛取最佳引物,并对反应体系和反应温度进行优化。结果表明,该方法采用20 μL体系在37 ℃恒温反应15 min即可对CP4-EPSPS基因进行快速检测。该方法检测转基因的灵敏度阈值为45拷贝,灵敏度高、特异性强,为大规模筛查CP4-EPSPS基因提供了一种新的途径。
中图分类号:
胡秀文, 邓波, 王金斌, 刘华, 唐雪明, 王宇, 曾海娟, 蒋玮, 李红. 基于RPA技术对转CP4-EPSPS基因产品的快速检测[J]. 中国农业科技导报, 2023, 25(2): 227-233.
Xiuwen HU, Bo DENG, Jinbin WANG, Hua LIU, Xueming TANG, Yu WANG, Haijuan ZENG, Wei JIANG, Hong LI. Rapid Detection of CP4-EPSPS Transgenic Products Based on RPA Technology[J]. Journal of Agricultural Science and Technology, 2023, 25(2): 227-233.
转基因作物 Genetically modified crop | 名称 Name | CP4-EPSPS基因 CP4-EPSPS gene |
|---|---|---|
| 转基因玉米 Transgenic maize | NK603 | 阳性 Positive |
| 转基因大豆 Transgenic Soybean | MON89788 | 阳性 Positive |
| 转基因棉花 Transgenic cotton | MON1445, MON88913 | 阳性 Positive |
| 转基因油菜 Transgenic rapeseed | GT73 | 阳性 Positive |
| 转基因苜蓿 Transgenic alfalfa | J101; J163 | 阳性 Positive |
| 转基因甜菜 Transgenic beet | H7-1 | 阳性 Positive |
| 转基因玉米 Transgenic maize | BT11; 59112; MON863 | 阴性 Negative |
| 转基因油菜 Transgenic rapeseed | RF1 | 阴性 Negative |
| 转基因水稻 Transgenic rice | KMD | 阴性 Negative |
表1 试验材料
Table 1 Experimental materials
转基因作物 Genetically modified crop | 名称 Name | CP4-EPSPS基因 CP4-EPSPS gene |
|---|---|---|
| 转基因玉米 Transgenic maize | NK603 | 阳性 Positive |
| 转基因大豆 Transgenic Soybean | MON89788 | 阳性 Positive |
| 转基因棉花 Transgenic cotton | MON1445, MON88913 | 阳性 Positive |
| 转基因油菜 Transgenic rapeseed | GT73 | 阳性 Positive |
| 转基因苜蓿 Transgenic alfalfa | J101; J163 | 阳性 Positive |
| 转基因甜菜 Transgenic beet | H7-1 | 阳性 Positive |
| 转基因玉米 Transgenic maize | BT11; 59112; MON863 | 阴性 Negative |
| 转基因油菜 Transgenic rapeseed | RF1 | 阴性 Negative |
| 转基因水稻 Transgenic rice | KMD | 阴性 Negative |
引物名称 Primer name | 引物序列 Primer sequences (5’-3’) | 扩增产物大小 Amplification product length/bp | GenBank ID |
|---|---|---|---|
| RPA-CP4-1-F | CCAATCACCTACAGGGTACCTATGGCTTCCGCTCA | 172 | KJ701163 |
| RPA-CP4-1-R | CAGCATCAGTCTCAACGGTAAGGTTAGCACCAAAA | ||
| RPA-CP4-2-F | CACTGAAAAGATGCTTCAAGGTTTTGGTGCTAACC | 150 | |
| RPA-CP4-2-R | AATGGGAAAGCAGTAGAGGATGGATCACCTGGAAC | ||
| RPA-CP4-3-F | TCAACACCCCAGGTATCACCACTGTTATCGAGCCA | 148 | |
| RPA-CP4-3-R | AGCTTACCACGACCTTCAAGACGGATGGTACGCAC | ||
| RPA-CP4-4-F | CAATCACCTACAGGGTACCTATGGCTTCCGCTC | 167 | |
| RPA-CP4-4-R | ATCAGTCTCAACGGTAAGGTTAGCACCAAAA | ||
| RPA-CP4-5-F | TCTCAACACCCCAGGTATCACCACTGTTATCGAGC | 204 | AF464188 |
| RPA-CP4-5-R | CAGCAACCAATGGGAAAGCAGTAGAGGATGGATCA |
表2 RPA引物
Table 2 RPA primers
引物名称 Primer name | 引物序列 Primer sequences (5’-3’) | 扩增产物大小 Amplification product length/bp | GenBank ID |
|---|---|---|---|
| RPA-CP4-1-F | CCAATCACCTACAGGGTACCTATGGCTTCCGCTCA | 172 | KJ701163 |
| RPA-CP4-1-R | CAGCATCAGTCTCAACGGTAAGGTTAGCACCAAAA | ||
| RPA-CP4-2-F | CACTGAAAAGATGCTTCAAGGTTTTGGTGCTAACC | 150 | |
| RPA-CP4-2-R | AATGGGAAAGCAGTAGAGGATGGATCACCTGGAAC | ||
| RPA-CP4-3-F | TCAACACCCCAGGTATCACCACTGTTATCGAGCCA | 148 | |
| RPA-CP4-3-R | AGCTTACCACGACCTTCAAGACGGATGGTACGCAC | ||
| RPA-CP4-4-F | CAATCACCTACAGGGTACCTATGGCTTCCGCTC | 167 | |
| RPA-CP4-4-R | ATCAGTCTCAACGGTAAGGTTAGCACCAAAA | ||
| RPA-CP4-5-F | TCTCAACACCCCAGGTATCACCACTGTTATCGAGC | 204 | AF464188 |
| RPA-CP4-5-R | CAGCAACCAATGGGAAAGCAGTAGAGGATGGATCA |
| 成分Ingredient | 反应体积 Reaction volume/μL | |||
|---|---|---|---|---|
| 50 | 25 | 20 | 15 | |
| Buffer | 29.50 | 14.75 | 11.80 | 8.85 |
| Primer-F | 2.40 | 1.20 | 1.00 | 0.70 |
| Primer-R | 2.40 | 1.20 | 1.00 | 0.70 |
| DNA | 2.00 | 1.00 | 0.80 | 0.60 |
| ddH2O | 11.20 | 5.60 | 4.20 | 3.40 |
| MgOAC | 2.50 | 1.25 | 1.00 | 0.75 |
表3 RPA体系优化
Table 3 RPA system optimization
| 成分Ingredient | 反应体积 Reaction volume/μL | |||
|---|---|---|---|---|
| 50 | 25 | 20 | 15 | |
| Buffer | 29.50 | 14.75 | 11.80 | 8.85 |
| Primer-F | 2.40 | 1.20 | 1.00 | 0.70 |
| Primer-R | 2.40 | 1.20 | 1.00 | 0.70 |
| DNA | 2.00 | 1.00 | 0.80 | 0.60 |
| ddH2O | 11.20 | 5.60 | 4.20 | 3.40 |
| MgOAC | 2.50 | 1.25 | 1.00 | 0.75 |
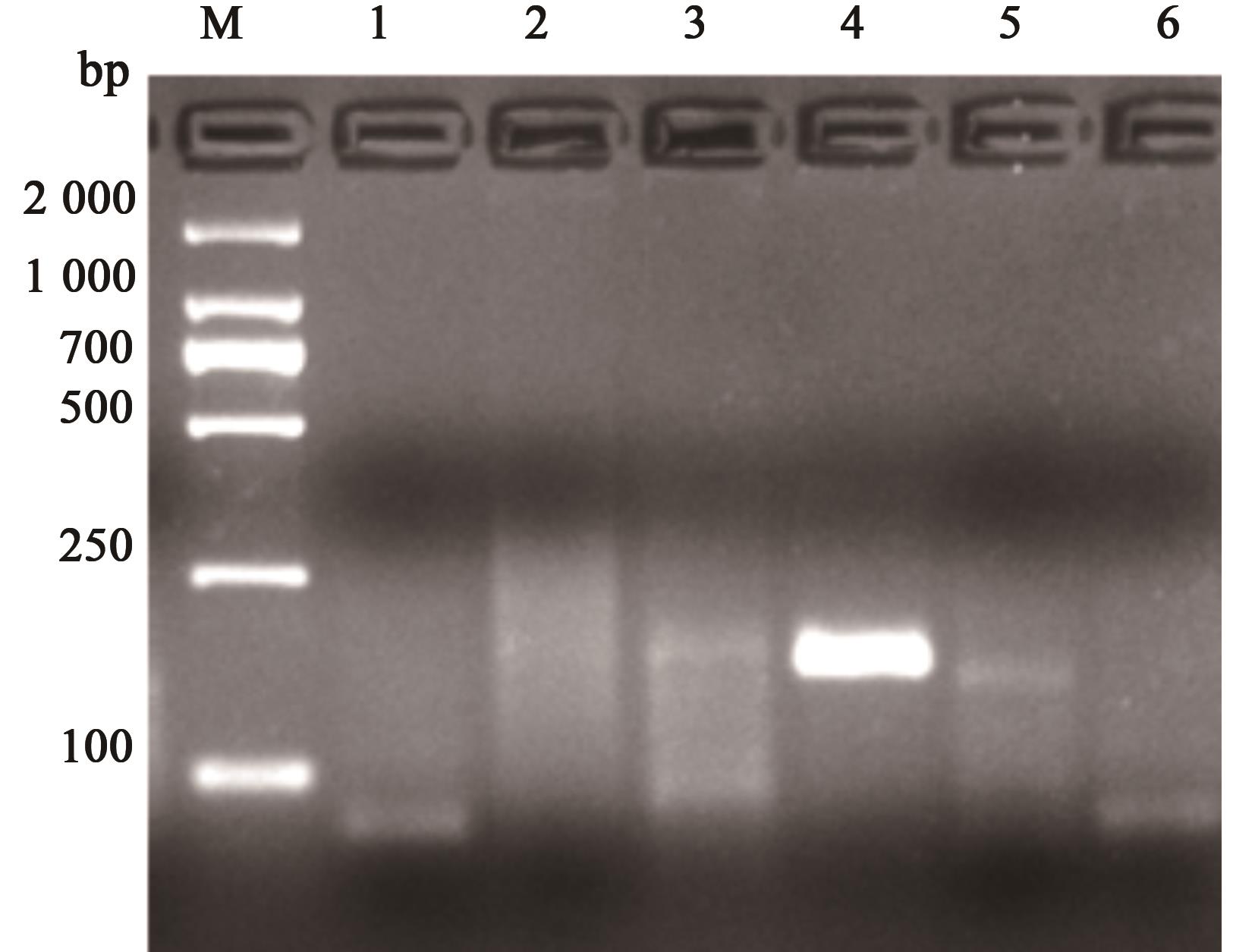
图1 RPA-CP4-1~RPA-CP4-5的扩增产物注:1~5分别为引物RPA-CP4-1~RPA-CP4-5;6为空白对照;M为2 000的maker。
Fig. 1 Amplification products of RPA-CP4-1~RPA-CP4-5Note:1~5 represent the amplified bands of RPA primers RPA-CP4-1~RPA-CP4-5, respectively; 6 represents the blank control; M is the maker of 2 000.
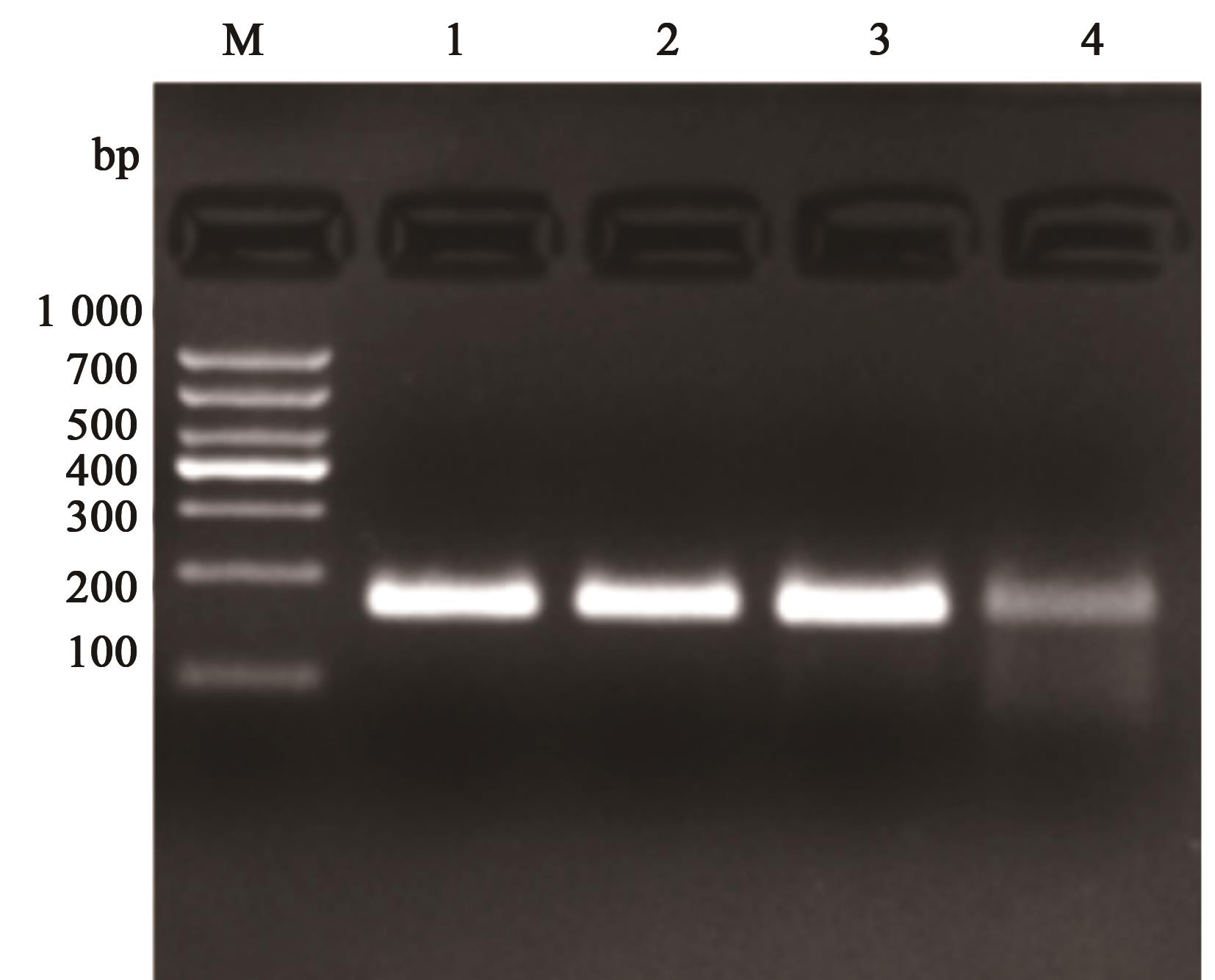
图2 不同体积反应体系的扩增产物注:1~4分别表示体系为50、25、20 和15 μL。
Fig. 2 Amplification products of different reaction systemNote:1~4 indicate that the system of 50, 25, 20 and 15 μL, respectively.
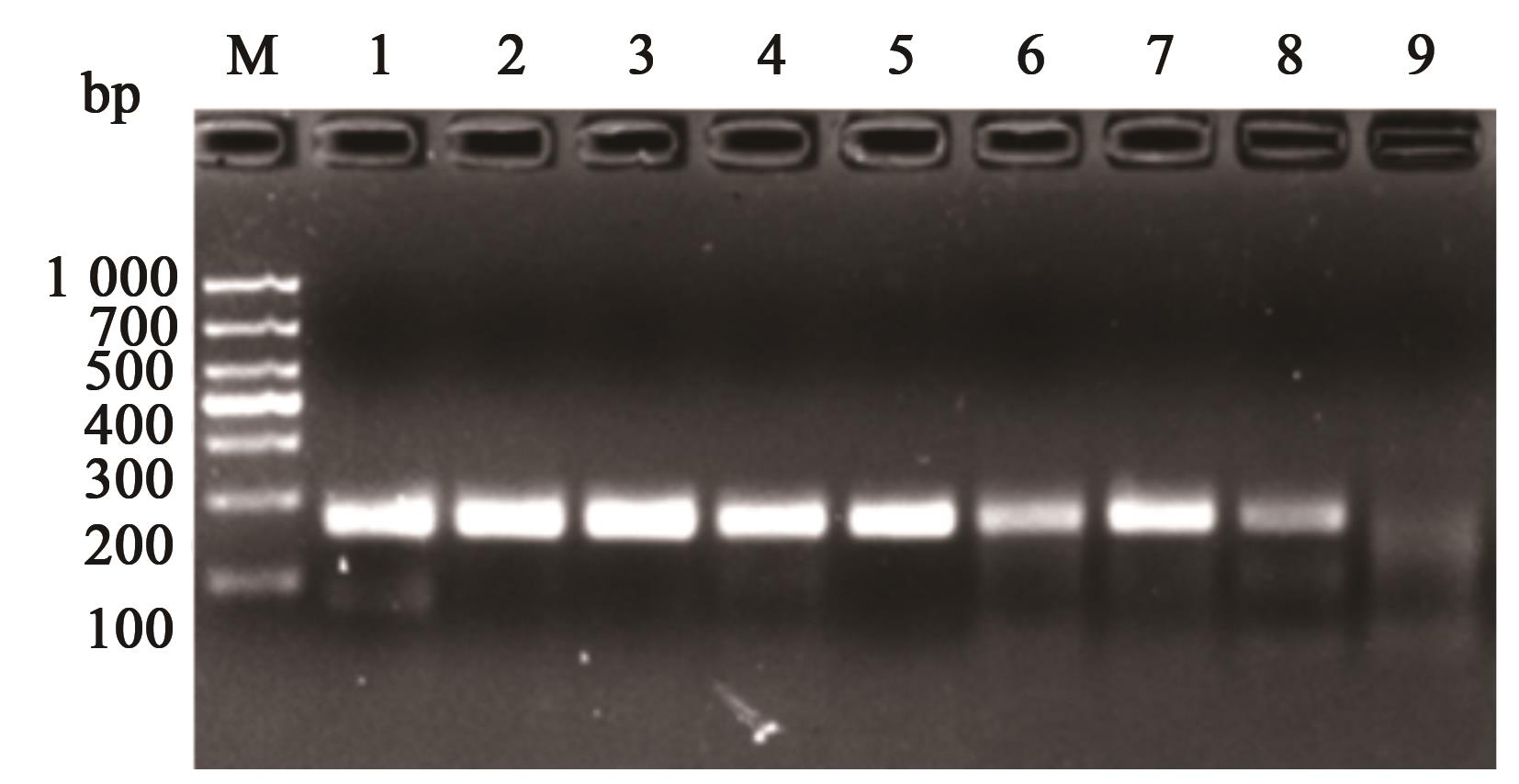
图3 不同反应温度的扩增产物注:1~8分别表示温度35、36、37、38、39、40、41、42 ℃;9空白对照;M为1 000的maker。
Fig. 3 Amplification products of different reaction temperaturesNote:1~8 represent the maker with temperature 35, 36, 37, 38, 39, 40, 41, 42 ℃; 9 is blank control, M is the maker of 1 000.
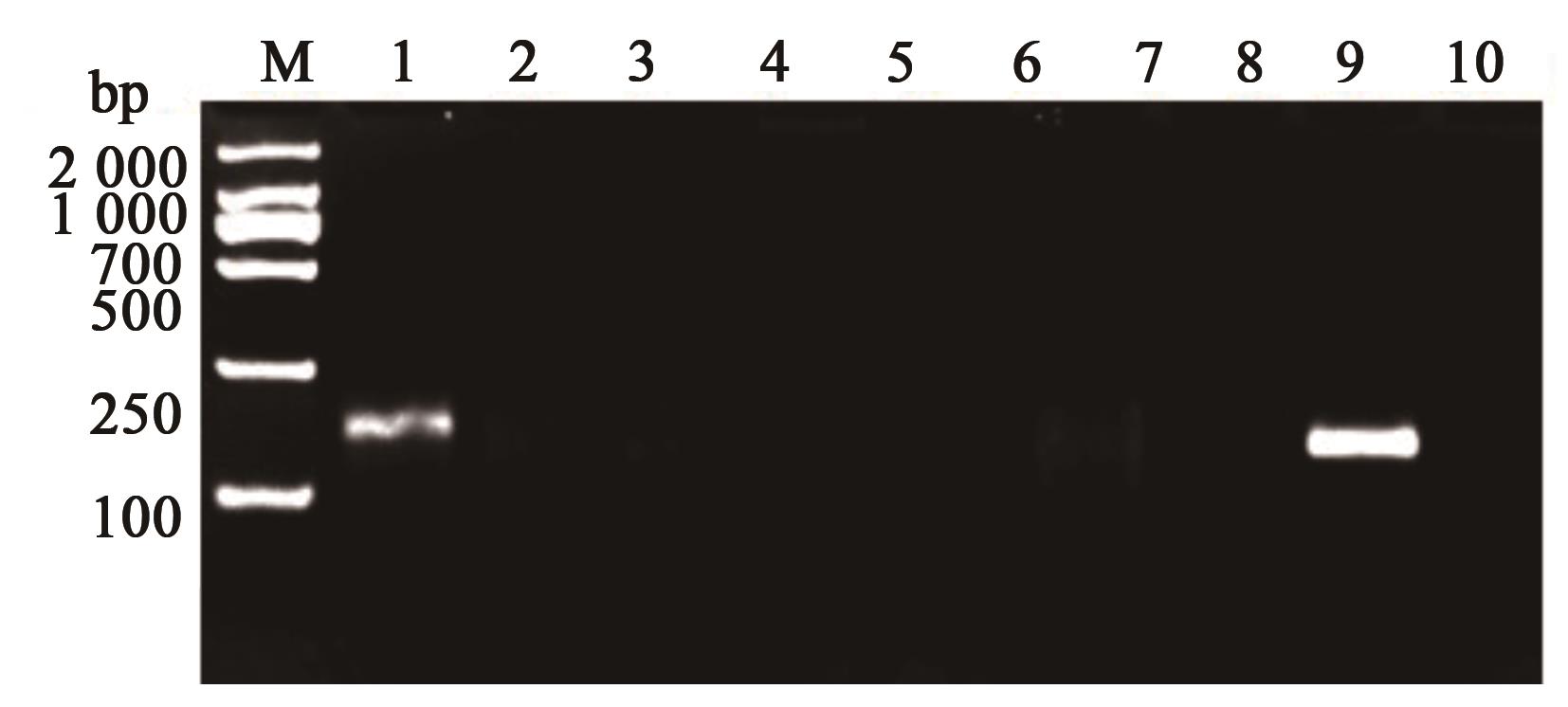
图4 RPA特异性检测注:1—2~9的DNA混合样品;2—转基因玉米BT-11;3—转基因水稻KMD;4—转基因玉米59112;5—转基因玉米MON863;6—转基因油菜RF1;7—非转基因大豆;8—非转基因玉米;9—转基因大豆MON88913。
Fig. 4 RPA specificity detectionNote:1—Mixture of 2~9; 2—Genetically modified corn BT-11; 3—Genetically modified rice KMD; 4—Genetically modified corn 59112; 5—Genetically modified corn MON863; 6—Genetically modified rape RF1; 7—Non-transgenic soybean; 8—Non-transgenic corn; 9—Genetically modified soybean MON88913.
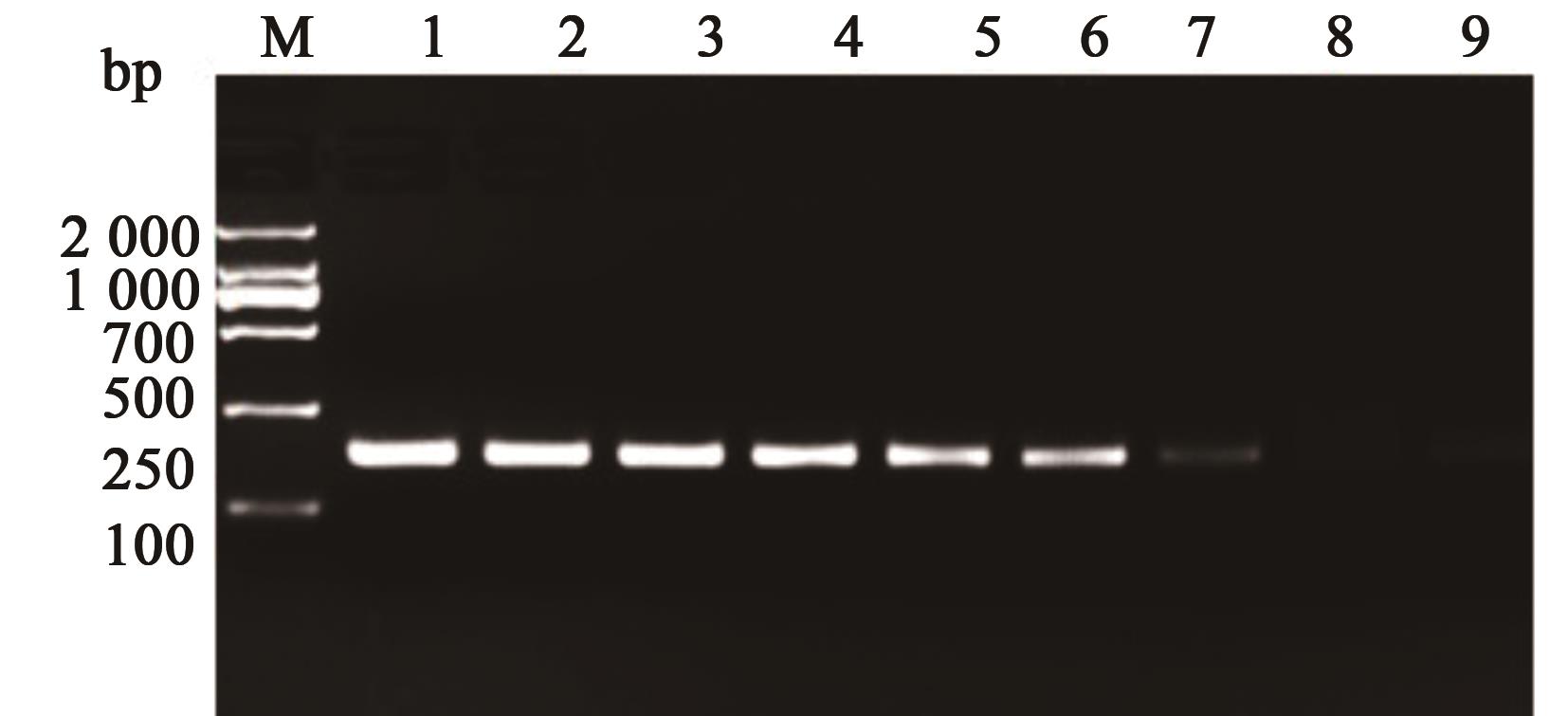
图5 RPA灵敏度检测注:1—4.5×107;2—4.5×106;3—4.5×105;4—4.5×104;5—4.5×103;6—4.5×102;7—4.5×101;8—4.5×100;9—空白对照。
Fig. 5 RPA sensitivity detectionNote:1—4.5×107; 2—4.5×106; 3—4.5×105; 4—4.5×104; 5—4.5×103; 6—4.5×102; 7—4.5×101; 8—4.5×100; 9—Blank control.
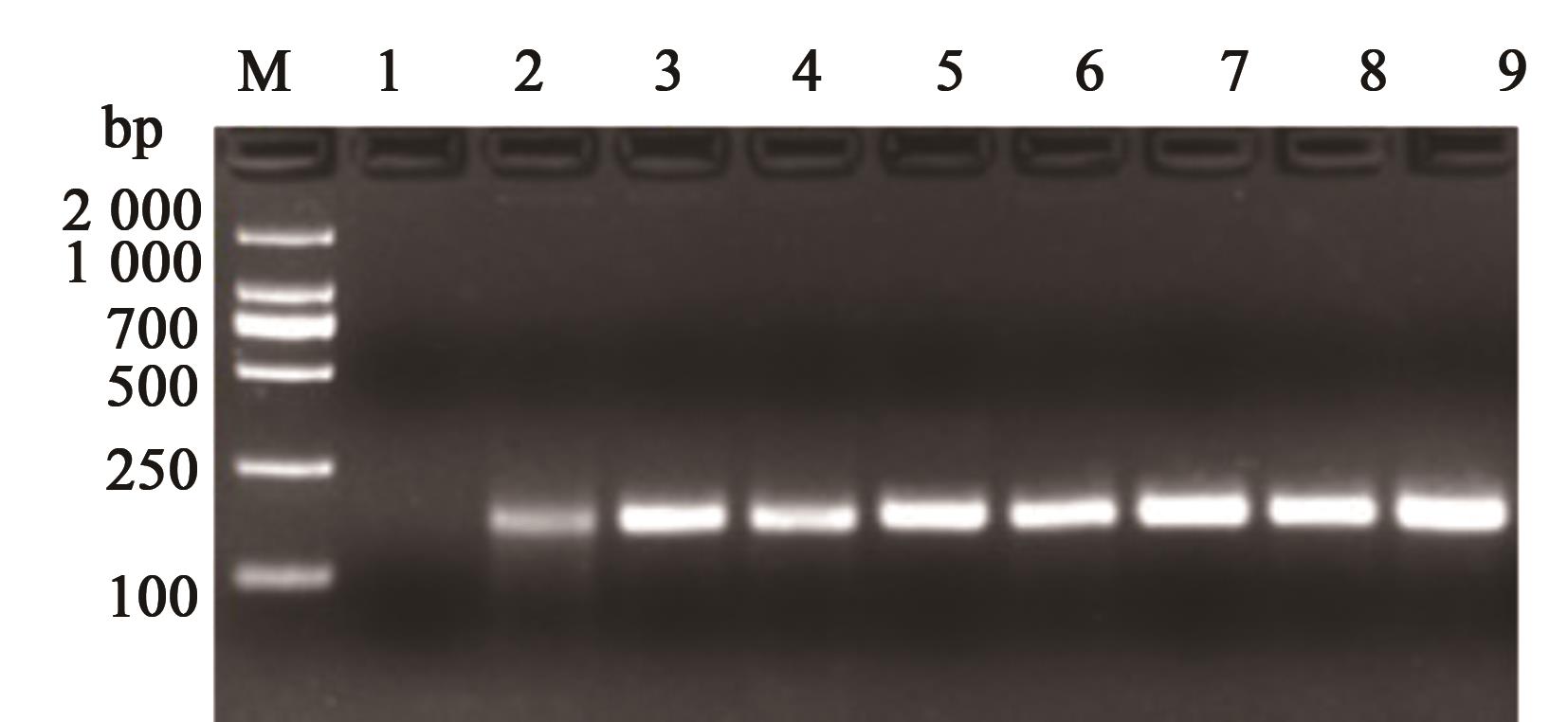
图6 转基因制品检测注:1—空白对照;2—转基因苜蓿J101;3—转基因甜菜H7-1;4—转基因油菜GT73;5—转基因棉花MON88913;6—转基因棉花MON1445;7—转基因苜蓿J163;8—转基因大豆MON89788;9—转基因玉米NK603。
Fig. 6 Detection of genetically modified productsNote:1—Blank control; 2—Transgenic alfalfa J101; 3—Transgenic sugar beet H7-1; 4—Transgenic rape GT73; 5—Transgenic cotton MON88913; 6—Transgenic cotton MON1445; 7—Transgenic alfalfa J163; 8—Transgenic Soybean MON89788; 9—Transgenic corn NK603.
分析方法 Analytical method | 检测靶标 Detection target | 灵敏度Sensitivity | 时间 Time/min | 使用条件 Conditions required | 参考文献Reference |
|---|---|---|---|---|---|
| 重组酶聚合酶扩增 RPA | CP4-EPSPS | 高High | 15 | Laboratory 实验室 | [ |
| 聚合酶链反应 PCR | PAT, BAR, Cry1Ab/Ac, p35s, tNOS, CTP2-EPSPS | 高High | 90 | Laboratory 实验室 | [ |
| 数字多聚酶链式反应 Digital PCR | MON87705, MON87769, DP356043 | 高High | 90 | Laboratory 实验室 | [ |
| 环介导等温扩增 LAMP | CP4-EPSPS | 高High | 60 | Laboratory 实验室 | [ |
| 酶联免疫反应 ELISA | Cry1 | 高High | 120 | Laboratory 实验室 | [ |
| 电化学免疫传感器Electrochemical immunosensor | CP4-EPSPS | 高High | 120 | Laboratory 实验室 | [ |
| 实时荧光定量 qPCR | CP4-EPSPS | 高High | 90 | Laboratory 实验室 | [ |
表4 核酸检测方法比较
Table 4 Comparison of nucleic acid detection methods
分析方法 Analytical method | 检测靶标 Detection target | 灵敏度Sensitivity | 时间 Time/min | 使用条件 Conditions required | 参考文献Reference |
|---|---|---|---|---|---|
| 重组酶聚合酶扩增 RPA | CP4-EPSPS | 高High | 15 | Laboratory 实验室 | [ |
| 聚合酶链反应 PCR | PAT, BAR, Cry1Ab/Ac, p35s, tNOS, CTP2-EPSPS | 高High | 90 | Laboratory 实验室 | [ |
| 数字多聚酶链式反应 Digital PCR | MON87705, MON87769, DP356043 | 高High | 90 | Laboratory 实验室 | [ |
| 环介导等温扩增 LAMP | CP4-EPSPS | 高High | 60 | Laboratory 实验室 | [ |
| 酶联免疫反应 ELISA | Cry1 | 高High | 120 | Laboratory 实验室 | [ |
| 电化学免疫传感器Electrochemical immunosensor | CP4-EPSPS | 高High | 120 | Laboratory 实验室 | [ |
| 实时荧光定量 qPCR | CP4-EPSPS | 高High | 90 | Laboratory 实验室 | [ |
| 1 | 国际农业生物技术应用服务组织.2019年全球生物技术/转基因作物商业化发展态势[J].中国生物工程杂志,2021,41(1):114-119. |
| 2 | 吴珊,庞俊琴,庄军红,等.我国转基因作物的研发与安全管理[J].中国农业科技导报,2020,22(11):11-16. |
| WU S, PANG J Q, ZHUANG J H, et al.. Research and development and safety management of genetically modified crops in China [J]. J. Agric. Sci. Technol., 2020, 22(11):11-16. | |
| 3 | MATHUR V, JAVID L, KULSHRESTHA S, et al.. World Cultivation of Genetically Modified Crops: Opportunities and Risks [M]. Sustainable Agriculture Reviews, Springer, 2017:45-87. |
| 4 | LIN C H, PAN T M. Perspectives on genetically modified crops and food detection [J]. J. Food. Drug. Anal., 2016, 24(1):1-8. |
| 5 | CASTAN M, ALI S, HOCHEGGER R, et al.. Analysis of the genetic stability of event NK603 in stacked corn varieties using high-resolution melting (HRM) analysis and Sanger sequencing [J]. Eur. Food Res. Technol., 2016, 243(3):1-13. |
| 6 | DILL G M, CAJACOB C A, PADGETTE S R. Glyphosate-resistant crops: adoption, use and future considerations [J]. Pest. Manag. Sci., 2008, 64(4):326-331. |
| 7 | KUMAR K, GAMBHIR G, DASS A, et al.. Genetically modified crops: current status and future prospects [J/OL]. Planta, 2020, 251(4):91 [2021-08-10]. . |
| 8 | 焦健.2020年上半年欧盟RASFF系统对华农产品通报分析[J].高科技与产业化,2020(9):72-75. |
| JIAO J. Analysis of EU RASFF notifications to China’s agricultural products in the first half of 2020 [J]. High-Technol. Ind., 2020(9):72-75. | |
| 9 | HOLST-JENSEN A. Testing for genetically modified organisms (GMOs): past, present and future perspectives [J]. Biotechnol. Adv., 2009, 27(6):1071-1082. |
| 10 | HADIDI A, KYRIAKOPOULOU P E, BARBA M. Major Advances in the History of Plant Virology [M]. New York: Academic Press, 2020:3-24. |
| 11 | RUBIO L, GALIPIENSO L, FERRIOL I. Detection of plant viruses and disease management: relevance of genetic diversity and evolution [J]. Front Plant Sci., 2020, 11:1092-1114. |
| 12 | CHEN Q, YAO C, YANG C, et al.. Development of an in-situ signal amplified electrochemical assay for detection of Listeria monocytogenes with label-free strategy [J]. Food Chem., 2021, 358:129894-129990. |
| 13 | PRASITPORN T, SENAPIN S, VANIKSAMPANNA A, et al.. Development of cross-priming amplification (CPA) combined with colorimetric and lateral flow dipstick visualization for scale drop disease virus (SDDV) detection [J]. J. Fish Dis., 2021. 44(9):1411-1422. |
| 14 | MARANO J M, CHUONG C, WEGER-LUCARELLI J. Rolling circle amplification: a high fidelity and efficient alternative to plasmid preparation for the rescue of infectious clones [J]. Virology, 2020, 551:58-63. |
| 15 | PIEPENBURG O, WILLIAMS C H, STEMPLE D L, et al.. DNA detection using recombination proteins [J]. Plos Biol., 2006, 4(7):1115-1121. |
| 16 | ZIA Q, ALAWAMI M, MOKHTAR N F K, et al.. Current analytical methods for porcine identification in meat and meat products [J]. Food Chem., 2020, 324:126664-126743. |
| 17 | DONG Y, ZHAO P P, CHEN L, et al.. Fast, simple and highly specific molecular detection of Vibrio alginolyticus pathogenic strains using a visualized isothermal amplification method [J]. BMC Vet. Res., 2020, 16(1):76-87. |
| 18 | LI J, MACDONALD J, STETTEN V. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification [J]. Analyst, 2020, 145(5):1950-1960. |
| 19 | 王亚楠,陈昌国.重组酶聚合酶扩增技术研究进展[J].解放军医学杂志,2021,46(5):504-511. |
| WANG Y N, CHEN C G. Research progress of recombinant polymerase amplification technology [J]. PLA Med. J., 2021, 46(5):504-511. | |
| 20 | GARRIDO-MAESTU A, AZINHEIRO S, CARVALHO J, et al.. Development and evaluation of loop-mediated isothermal amplification, and recombinase polymerase amplification methodologies, for the detection of listeria monocytogenes in ready to-eat food samples [J]. Food Control, 2018, 86:27-34. |
| 21 | MCQUILLAN J S, WILSON M W. Recombinase polymerase amplification for fast, selective, DNA-based detection of faecal indicator Escherichia coli [J]. Lett. App. Microbiol., 2021, 72(4):382-389. |
| 22 | PANG Y, CONG F, ZHANG X, et al.. A recombinase polymerase amplification-based assay for rapid detection of Chlamydia psittaci [J]. Poult. Sci., 2021, 100(2):585-591. |
| 23 | WAND N I V, BONNEY L C, Watson R J, et al.. Point-of-care diagnostic assay for the detection of Zika virus using the recombinase polymerase amplification method [J]. J. Gen. Virol., 2018, 99(8):1012-1026. |
| 24 | WANG X, XIE S, CHEN X, et al.. A rapid and convenient method for on-site detection of MON863 maize through real-time fluorescence recombinase polymerase amplification [J]. Food Chem., 2020, 324:126821-126826. |
| 25 | 龙丽坤,赵宁,夏蔚,等.转基因玉米CM8101特异性定性PCR检测方法[J].中国农学通报,2021,37(23):23-28. |
| LONG L K, ZHAO N, XIA W, et al.. Detection of CM8101 specific qualitative PCR in transgenic maize [J]. Chin. Agric. Sci. Bull., 2021, 37(23):23-28. | |
| 26 | 卢海强,焦新雅,吴思源,等.数字PCR在食源性致病菌检测中的应用进展[J].生物技术进展,2021,11(3):260-268. |
| LU H Q, JIAO X Y, WU S Y, et al.. Application progress of digital PCR in detection of foodborne pathogens [J]. Progress Biotechnol., 2021, 11(3): 260-268. | |
| 27 | 单大鹏, 王晓云, 洪志鹏, 等. 转Cry1Ia基因抗虫大豆对鳞翅目靶标害虫的抗性分析[J].东北农业大学学报,2021,52(5): 1-9. |
| SHAN D P, WANG X Y, HONG Z P, et al.. Analysis of the resistance of transgenic Cry1Ia insect-resistant soybean to lepidopteran target pests [J]. J. Northeast Agric. Univ., 2021, 52(5):1-9. | |
| 28 | 高鸿飞. 基于免疫传感新方法的外源蛋白分析研究[D].武汉:华中农业大学,2020. |
| GAO H F. Research on the analysis of foreign proteins based on new methods of immunosensing [D]. Wuhan: Huazhong Agricultural University, 2020. |
| [1] | 李浩辉, 刘彩月, 张海文, 王旭静, 唐巧玲, 王友华. 2022年度全球转基因作物产业化发展现状及趋势分析[J]. 中国农业科技导报, 2023, 25(12): 6-16. |
| [2] | 吴珊,庞俊琴,庄军红,陈丽梅*. 我国转基因作物的研发与安全管理[J]. 中国农业科技导报, 2020, 22(11): 11-16. |
| [3] | 安娜1,2,郑子繁2,董美2,刘卫晓2,宛煜嵩2,金芜军2,李亮2*,韩阳1*. 基于表面等离子体共振的核酸传感器研究进展[J]. 中国农业科技导报, 2019, 21(5): 55-61. |
| [4] | 孙卓婧1,张安红2,叶纪明1*. 转基因作物研发现状及展望[J]. 中国农业科技导报, 2018, 20(7): 11-18. |
| [5] | 张立兰,陈亮,张宏福*. 转基因作物对畜禽肠道非预期效应的研究进展[J]. 中国农业科技导报, 2018, 20(11): 1-13. |
| [6] | 祁潇哲,黄昆仑*. 转基因食品安全评价研究进展[J]. , 2013, 15(4): 14-19. |
| [7] | 李鑫鑫1,李晓晖1,李亮1,平淑珍1,陈明1,张维1,燕永亮1,赵新宇2,陆伟1. 转基因作物对于土壤微生物的影响[J]. , 2010, 12(6): 24-27. |
| [8] | 周长发,张锐,张晓,罗淑萍,郭三堆. 地高辛随机引物法标记探针的Southern杂交技术优化[J]. , 2009, 11(4): 123-128. |
| [9] | 徐鸿林,翟红利,王锋,朴建华,杨晓光,朱祯. 豇豆胰蛋白酶抑制剂基因(cpti)及其在抗虫转基因作物中的应用[J]. , 2008, 10(1): 18-27. |
| [10] | 刘蓉蓉|文学. 遗传利用限制技术(GURTs)的发展及应用前景[J]. , 2008, 10(1): 58-62. |
| [11] | Valerie J. Karplus[1] 邓兴旺[1,2,3]. 从实验室到田间:中国农业生物技术的发展及其影响[J]. , 2007, 9(3): 4-8. |
| [12] | 陈洁君[1] 王劲[2] 宛煜嵩[3] 金芜军[3]. 转基因作物安全性评价与商品化前景分析[J]. , 2007, 9(3): 38-43. |
| [13] | 张军民[1] 胡广东[2] 等. 转基因食品与饲料安全及其评价[J]. , 2002, 4(4): 21-26. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||