




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (12): 30-38.DOI: 10.13304/j.nykjdb.2024.0156
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Zhan ZHAO1( ), Xiaoting WANG1(
), Xiaoting WANG1( ), Lifeng ZHANG2, Jinhe ZHAO2, Yuhong YU2, Junhua LI2, Zhanqing WU2
), Lifeng ZHANG2, Jinhe ZHAO2, Yuhong YU2, Junhua LI2, Zhanqing WU2
Received:2024-03-04
Accepted:2024-07-13
Online:2024-12-15
Published:2024-12-17
Contact:
Xiaoting WANG
赵展1( ), 王晓婷1(
), 王晓婷1( ), 张黎凤2, 赵津禾2, 于玉红2, 李军华2, 吴占清2
), 张黎凤2, 赵津禾2, 于玉红2, 李军华2, 吴占清2
通讯作者:
王晓婷
作者简介:赵展 E-mail:13525579383@163.com;
基金资助:CLC Number:
Zhan ZHAO, Xiaoting WANG, Lifeng ZHANG, Jinhe ZHAO, Yuhong YU, Junhua LI, Zhanqing WU. Transcriptome Analysis of Watermelon Responses to Low Nitrogen Stress[J]. Journal of Agricultural Science and Technology, 2024, 26(12): 30-38.
赵展, 王晓婷, 张黎凤, 赵津禾, 于玉红, 李军华, 吴占清. 西瓜对低氮胁迫响应的转录组分析[J]. 中国农业科技导报, 2024, 26(12): 30-38.
Add to citation manager EndNote|Ris|BibTeX
URL: https://nkdb.magtechjournal.com/EN/10.13304/j.nykjdb.2024.0156
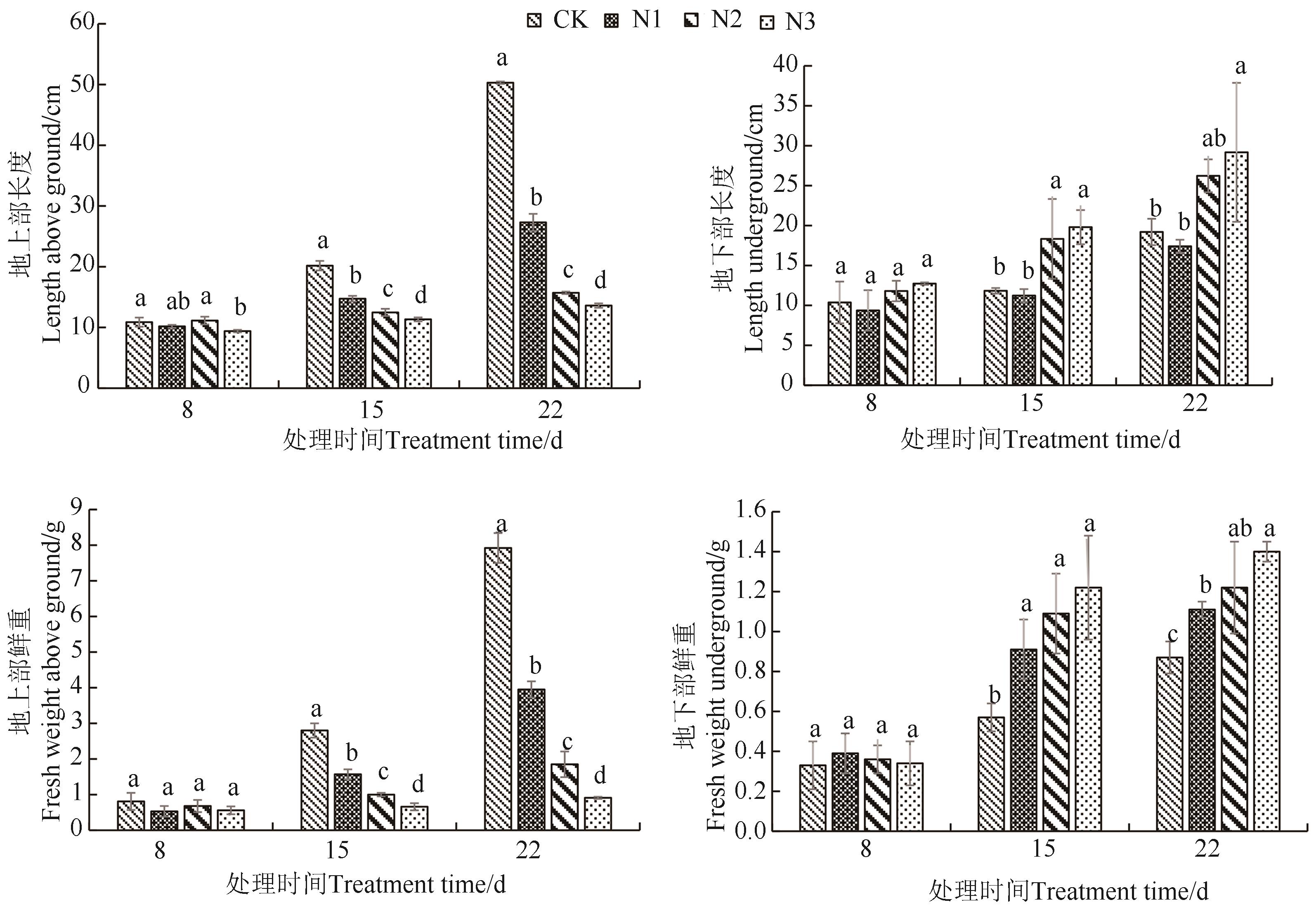
Fig. 1 Phenotype of watermelon seedling under different treatmentsNote:Different lowercase letters indicate significant differences between different treatments of same time at P<0.05 level.
样品 Sample | 有效数据 Clean reads | 比对数据 Mapped reads | 比对率 Mapping ratio/% | Q30/% | GC含量 GC content/% |
|---|---|---|---|---|---|
| CK | 44 680 604 | 43 215 897 | 96.73 | 91.57 | 45.52 |
| N1 | 42 755 765 | 41 534 388 | 97.16 | 91.05 | 45.98 |
| N3 | 44 968 308 | 43 682 840 | 97.14 | 91.43 | 44.94 |
Table 1 Transcriptome sequencing data quality statistics of watermelon in different N contents
样品 Sample | 有效数据 Clean reads | 比对数据 Mapped reads | 比对率 Mapping ratio/% | Q30/% | GC含量 GC content/% |
|---|---|---|---|---|---|
| CK | 44 680 604 | 43 215 897 | 96.73 | 91.57 | 45.52 |
| N1 | 42 755 765 | 41 534 388 | 97.16 | 91.05 | 45.98 |
| N3 | 44 968 308 | 43 682 840 | 97.14 | 91.43 | 44.94 |
样品 Sample | 差异表达基因数量 Number of DEGs | ||
|---|---|---|---|
总计 Total | 上调 Up-regulated | 下调 Down-regulated | |
| CK/N1 | 121 | 34 | 87 |
| N1/N3 | 1 184 | 843 | 341 |
| CK/N3 | 516 | 327 | 189 |
Table 2 DEGs among treatments
样品 Sample | 差异表达基因数量 Number of DEGs | ||
|---|---|---|---|
总计 Total | 上调 Up-regulated | 下调 Down-regulated | |
| CK/N1 | 121 | 34 | 87 |
| N1/N3 | 1 184 | 843 | 341 |
| CK/N3 | 516 | 327 | 189 |
基因ID Gene ID | 调控 regulated | 基因注释 Gene annotation | Pfam注释 Pfam_annotation |
|---|---|---|---|
| Cla97C01G023010 | 下调Down | 类Fe2+转运蛋白 Fe2+ transport protein 1-like | ZIP 锌转运体 ZIP Zinc transporter |
| Cla97C04G071110 | 下调Down | 类ORG2 转录因子 Transcription factor ORG2-like | HLH的DNA结合域 Helix-loop-helix DNA-binding domain |
| Cla97C07G136330 | 下调Down | 琥珀酸脱氢酶[泛醌]铁硫亚基3 Succinate dehydrogenase [ubiquinone] iron-sulfur subunit 3 | 铁硫簇结合域 Iron-sulfur cluster binding domain; |
| Cla97C03G057100 | 上调 Up | 酪氨酸酶/多酚氧化酶 Polyphenol oxidase | 酪氨酸酶的共同中心结构域,多酚氧化酶中间结构域 Common central domain of tyrosinase, polyphenol oxidase middle domain |
| Cla97C08G150960 | 下调Down | 海马丰富的转录样蛋白 Hippocampus abundant transcript-like protein 1 | MFS/糖转运蛋白 MFS/sugar transport protein |
| Cla97C02G040720 | 下调Down | 烟酰胺合成酶1 Nicotianamine synthase 1 | 烟胺合成酶蛋白 Nicotianamine synthase protein |
| Cla97C02G034900 | 下调Down | 推测蛋白Csa_2G423710 Hypothetical protein Csa_2G423710 | — |
| Cla97C06G124190 | 下调Down | 预测蛋白LOC101215023 Uncharacterized protein LOC101215023 | 氰菊酯HHE阳离子结合域,锌带,CHY锌指 Hemerythrin HHE cation binding domain, zinc-ribbon, CHY zinc finger |
| Cla97C02G034910 | 下调Down | 类Nramp3 金属转运体 Metal transporter Nramp3-like | 天然抗性相关巨噬蛋白 Natural resistance-associated macrophage protein |
| Cla97C04G071130 | 下调Down | bHLH100转录因子 Transcription factor bHLH100-like | HLH的DNA结合域 Helix-loop-helix DNA-binding domain |
| Cla97C01G021100 | 下调Down | 预测蛋白 Uncharacterized protein | 未知功能的类PDDEXK家族 PDDEXK-like family of unknown function |
| Cla97C05G091150 | 下调Down | 质体脂相关蛋白,叶绿体异构体X1 Probable plastid-lipid-associated protein 14, chloroplastic isoform X1 | 蛋白激酶结构域,PAP原纤蛋白 Protein kinase domain, PAP_fibrillin |
Table 3 DEGs and functional annotation between CK and N1
基因ID Gene ID | 调控 regulated | 基因注释 Gene annotation | Pfam注释 Pfam_annotation |
|---|---|---|---|
| Cla97C01G023010 | 下调Down | 类Fe2+转运蛋白 Fe2+ transport protein 1-like | ZIP 锌转运体 ZIP Zinc transporter |
| Cla97C04G071110 | 下调Down | 类ORG2 转录因子 Transcription factor ORG2-like | HLH的DNA结合域 Helix-loop-helix DNA-binding domain |
| Cla97C07G136330 | 下调Down | 琥珀酸脱氢酶[泛醌]铁硫亚基3 Succinate dehydrogenase [ubiquinone] iron-sulfur subunit 3 | 铁硫簇结合域 Iron-sulfur cluster binding domain; |
| Cla97C03G057100 | 上调 Up | 酪氨酸酶/多酚氧化酶 Polyphenol oxidase | 酪氨酸酶的共同中心结构域,多酚氧化酶中间结构域 Common central domain of tyrosinase, polyphenol oxidase middle domain |
| Cla97C08G150960 | 下调Down | 海马丰富的转录样蛋白 Hippocampus abundant transcript-like protein 1 | MFS/糖转运蛋白 MFS/sugar transport protein |
| Cla97C02G040720 | 下调Down | 烟酰胺合成酶1 Nicotianamine synthase 1 | 烟胺合成酶蛋白 Nicotianamine synthase protein |
| Cla97C02G034900 | 下调Down | 推测蛋白Csa_2G423710 Hypothetical protein Csa_2G423710 | — |
| Cla97C06G124190 | 下调Down | 预测蛋白LOC101215023 Uncharacterized protein LOC101215023 | 氰菊酯HHE阳离子结合域,锌带,CHY锌指 Hemerythrin HHE cation binding domain, zinc-ribbon, CHY zinc finger |
| Cla97C02G034910 | 下调Down | 类Nramp3 金属转运体 Metal transporter Nramp3-like | 天然抗性相关巨噬蛋白 Natural resistance-associated macrophage protein |
| Cla97C04G071130 | 下调Down | bHLH100转录因子 Transcription factor bHLH100-like | HLH的DNA结合域 Helix-loop-helix DNA-binding domain |
| Cla97C01G021100 | 下调Down | 预测蛋白 Uncharacterized protein | 未知功能的类PDDEXK家族 PDDEXK-like family of unknown function |
| Cla97C05G091150 | 下调Down | 质体脂相关蛋白,叶绿体异构体X1 Probable plastid-lipid-associated protein 14, chloroplastic isoform X1 | 蛋白激酶结构域,PAP原纤蛋白 Protein kinase domain, PAP_fibrillin |
| 1 | DU Q G, YANG J, SYED MUHAMMAD SADIQ S, et al.. Comparative transcriptome analysis of different nitrogen responses in low-nitrogen sensitive and tolerant maize genotypes [J]. J. Integr. Agric., 2021,20(8):2043-2055. |
| 2 | SHIN S Y, JEONG J S, LIM J Y, et al.. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies [J]. BMC Genom., 2018,19(1):1-20. |
| 3 | PENG S B, BURESH R J, HUANG J L, et al.. Improving nitrogen fertilization in rice by sitespecific N management.a review [J]. Agron. Sustain. Dev., 2010,30(3):649-656. |
| 4 | YANG F, ZHANG Z L, BARBERÁN A, et al.. Nitrogen-induced acidification plays a vital role driving ecosystem functions:insights from a 6-year nitrogen enrichment experiment in a Tibetan alpine meadow [J/OL].Soil Biol. Biochem., 2021,153:108107 [2024-02-20]. . |
| 5 | GAUDINIER A, RODRIGUEZ-MEDINA J, ZHANG L F, et al.. Transcriptional regulation of nitrogen-associated metabolism and growth [J]. Nature, 2018,563:259-264. |
| 6 | SHI X L, CUI F, HAN X Y, et al.. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204 [J]. Mol. Plant, 2022,15(9):1440-1456. |
| 7 | ZHOU Z H, WANG C K, ZHENG M H, et al.. Patterns and mechanisms of responses by soil microbial communities to nitrogen addition [J]. Soil Biol. Biochem., 2017,115:433-441. |
| 8 | LUO L, RAN L, RASOOL Q Z, et al.. Integrated modeling of U.S. agricultural soil emissions of reactive nitrogen and associated impacts on air pollution,health,and climate [J]. Environ. Sci. Technol., 2022,56(13):9265-9276. |
| 9 | LIU Q, CHEN X B, WU K, et al.. Nitrogen signaling and use efficiency in plants:what’s new? [J]. Curr.Opin.Plant Biol., 2015,27:192-198. |
| 10 | WANG G, WANG J, YAO L, et al.. Transcriptome and metabolome reveal the molecular mechanism of barley genotypes underlying the response to low nitrogen and resupply [J/OL]. Int. J. Mol. Sci., 2023,24(5):4706 [2024-02-20]. . |
| 11 | SCHEURWATER I, KOREN M, LAMBERS H, et al.. The contribution of roots and shoots to whole plant nitrate reduction in fast‐and slow‐growing grass species [J]. J.Exp.Bot., 2002,53(374):1635-1642. |
| 12 | MIFLIN B J. Distribution of nitrate and nitrite reductase in barley [J]. Nature, 1967,214:1133-1134. |
| 13 | NAWAZ M A, WANG L M, JIAO Y Y, et al.. Pumpkin rootstock improves nitrogen use efficiency of watermelon scion by enhancing nutrient uptake,cytokinin content,and expression of nitrate reductase genes [J]. Plant Growth Regul., 2017,82(2):233-246. |
| 14 | LUO J, LI H, LIU T X, et al.. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability [J]. J. Exp. Bot., 2013,64(14):4207-4224. |
| 15 | SAENGWILAI P, TIAN X L, LYNCH J P. Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize [J]. Plant Physiol., 2014,166(2):581-589. |
| 16 | WEI S S, WANG X Y, SHI D Y, et al.. The mechanisms of low nitrogen induced weakened photosynthesis in summer maize (Zea mays L.) under field conditions [J]. Plant Physiol.Biochem., 2016,105:118-128. |
| 17 | HE L, TENG L, TANG X,et al.. Agro-morphological and metabolomics analysis of low nitrogen stress response in Axonopus compressus [J/OL]. Aob Plants, 2021,13(4):plab022 [2024-02-20] . . |
| 18 | 董玥,陈雪平,赵建军,等.低氮胁迫不同氮效率基因型茄子光合特性差异[J].华北农学报,2009,24(1):181-184. |
| DONG Y, CHEN X P, ZHAO J J, et al.. Differences of photosynthetic characteristics in the nitrogen efficiency genotypes of eggplant under low nitrogen stress [J]. Acta Agric. Boreali-Sin., 2009,24(1):181-184. | |
| 19 | 李强,罗延宏,余东海,等.低氮胁迫对耐低氮玉米品种苗期光合及叶绿素荧光特性的影响[J].植物营养与肥料学报,2015,21(5):1132-1141. |
| LI Q, LUO Y H, YU D H, et al.. Effects of low nitrogen stress on photosynthetic characteristics and chlorophyll fluorescence parameters of maize cultivars tolerant to low nitrogen stress at the seedling stage [J]. J. Plant Nutr. Fert., 2015,21(5):1132-1141. | |
| 20 | LI L, GAO W, PENG Q,et al.. Two soybean bHLH factors regulate response to iron deficiency [J]. J.Integr.Plant Biol.,2018,60(7):608-622. |
| 21 | ZHANG C, HOU Y, HAO Q, et al.. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress [J/OL]. PLoS One,2015,10(4):e0125174 [2024-02-20]. . |
| 22 | QU B Y, HE X, WANG J, et al.. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input [J]. Plant Physiol., 2015,167(2):411-423. |
| 23 | TANG Y T, LI X, LU W, et al.. Enhanced photorespiration in transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase gene contributes to alleviating low nitrogen stress [J]. Plant Physiol. Biochem., 2018,130:577-588. |
| 24 | 肖燕,姚珺玥,刘冬,等.甘蓝型油菜响应低氮胁迫的表达谱分析[J].作物学报,2020,46(10):1526-1538. |
| XIAO Y, YAO J Y, LIU D, et al.. Expression profile analysis of low nitrogen stress in Brassica napus [J]. Acta Agron. Sin., 2020,46(10):1526-1538. | |
| 25 | YAN H, SHI H, HU C, et al.. Transcriptome differences in response mechanisms to low-nitrogen stress in two wheat varieties [J/OL]. Int. J. Mol. Sci., 2021,22(22):12278 [2024-02-20]. . |
| 26 | WANG W, XIN W, CHEN N,et al.. Transcriptome and co-expression network analysis reveals the molecular mechanism of rice root systems in response to low-nitrogen conditions [J/OL].Int. J. Mol. Sci., 2023,24(6):5290 [2024-02-20]. . |
| 27 | ALKHADER A M F, QARYOUTI M M, OKASHA T M. Effect of nitrogen on yield, quality, and irrigation water use efficiency of drip fertigated grafted watermelon (Citrullus lanatus) grown on a calcareous soil [J]. J. Plant Nutr., 2019,42(2):1-12. |
| 28 | 杜少平,马忠明,唐超男,等. 基于SPAD的西瓜氮素营养诊断与推荐施肥技术研究 [J]. 中国土壤与肥料, 2023 (12):200-209. |
| DU S P, MA Z M, TANG C N, et al.. Study on nitrogen nutrition diagnosis and fertilizer recommendation of watermelon based on SPAD value [J]. Soil Fert. Sci. China, 2023 (12):200-209. | |
| 29 | 张婷婷,孟丽丽,刘晓蕊,等.马铃薯氮代谢对低氮胁迫的响应及转录组分析[J].西北农林科技大学学报(自然科学版),2022,50(8):15-26. |
| ZHANG T T, MENG L L, LIU X R,et al.. Response of nitrogen metabolism of potato to low nitrogen stress and transcriptome analysis [J]. J. Northwest A&F Univ. (Nat. Sci.), 2022,50(8):15-26. | |
| 30 | HE X, QU B Y, LI W J,et al.. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield [J]. Plant Physiol.,2015,169(3):1991-2005. |
| 31 | WANG Y, ZHAO Y, WANG S,et al.. Up-regulated 2-alkenal reductase expression improves low-nitrogen tolerance in maize by alleviating oxidative stress [J]. Plant Cell Environ.,2021,44(2):559-573. |
| [1] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [2] | Lanting XIANG, Shuhui SONG, Lijuan LIU, Xiaoling SHE, Jiahua ZHOU, Baogang WANG, Hong CHANG, Chao ZHANG, Daqi FU, Yunxiang WANG. Effect of Different Storage Temperatures on Quality of Jingcai 1 Watermelon [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 137-145. |
| [3] | Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43. |
| [4] | Lin ZHANG, Yantao YANG, Lili SONG, Shiping MAO. Current Situation and High Quality Development Countermeasures of Watermelon and Muskmelon Industry in Beijing [J]. Journal of Agricultural Science and Technology, 2023, 25(11): 20-27. |
| [5] | ZHANG Wenyun1, ZHANG Jiancheng2, YAO Jingzhen2*. Comparative Transcriptome Analysis of Wheat Leaf in Response to Low Nitrogen Stress#br# [J]. Journal of Agricultural Science and Technology, 2020, 22(11): 26-34. |
| [6] | LI Peng-fei, HUO Xiu-ai, CHENG Yong-qiang, DAI Liang, YANG Bing-yan, DUAN Hui-ju. Assessment of Genetic Diversity in Watermelon Based on SRAP Analysis [J]. , 2013, 15(2): 89-96. |
| [7] | Gao Dong1, ZHOU Hong-pei2. The Discussion of Planting Density of Sweet Potato Intercropped with Watermelon in Greenhouse [J]. , 2009, 11(S2): 63-65. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号