




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (11): 226-239.DOI: 10.13304/j.nykjdb.2025.0240
• 方法与技术创新 • 上一篇
谢浩1,2( ), 陈指龙2,3,4,5, 彭翠婷2,3, 潘颖婷2,3, 齐霖2,3, 赵玉兰2,3, 闭英连6, 宋子仪1(
), 陈指龙2,3,4,5, 彭翠婷2,3, 潘颖婷2,3, 齐霖2,3, 赵玉兰2,3, 闭英连6, 宋子仪1( ), 唐中林2,3,4,5(
), 唐中林2,3,4,5( )
)
收稿日期:2025-04-04
接受日期:2025-05-26
出版日期:2025-11-15
发布日期:2025-11-17
通讯作者:
宋子仪,唐中林
作者简介:谢浩 E-mail:xiehao2024888@163.com
基金资助:
Hao XIE1,2( ), Zhilong CHEN2,3,4,5, Cuiting PENG2,3, Yingting PAN2,3, Lin QI2,3, Yulan ZHAO2,3, Yinglian BI6, Ziyi SONG1(
), Zhilong CHEN2,3,4,5, Cuiting PENG2,3, Yingting PAN2,3, Lin QI2,3, Yulan ZHAO2,3, Yinglian BI6, Ziyi SONG1( ), Zhonglin TANG2,3,4,5(
), Zhonglin TANG2,3,4,5( )
)
Received:2025-04-04
Accepted:2025-05-26
Online:2025-11-15
Published:2025-11-17
Contact:
Ziyi SONG,Zhonglin TANG
摘要:
CRISPR/Cas9作为一款强大的基因编辑工具,在猪的遗传改良中已经广泛应用。然而,单一基因的编辑无法满足多性状的同步改良。为利用CRISPR/Cas9基因编辑体系构建MSTN、pAPN和CD163多基因编辑猪胚胎成纤维细胞(porcine embryonic fibroblasts, PEF)并制备胚胎,首先,根据猪CD163、MSTN和pAPN蛋白功能结构域,选定MSTN外显子1、CD163外显子7及pAPN外显子2作为打靶区域,各设计2对sgRNAs,连接至骨架pX330载体,通过电转染至PEF,筛选每个基因编辑效率较高的sgRNA;然后,利用同源重组法将CD163、MSTN、pAPN单个基因的sgRNA表达盒组装成一体化串联表达系统,转化至Fast-T1感受态细胞并进行Sanger测序鉴定;最后,利用电转染方式将3基因敲除载体转染至PEF,用2.5 μg·mL-1的嘌呤霉素进行药筛,挑选单克隆细胞,PCR扩增后进行Sanger测序,鉴定单克隆细胞MSTN、CD163、pAPN基因的打靶序列。结果显示,CD163、MSTN、pAPN基因的sgRNA1编辑效率高于sgRNA2,因此选择较高效率sgRNA1用于构建一体化质粒。质粒测序显示,3个基因的sgRNA表达盒成功连接到骨架pX330载体上。挑取45株转染3基因编辑质粒的单克隆细胞,测序结果显示,MSTN、pAPN、CD163基因的突变率分别为62%、26%、11%,其中有4株细胞实现了3个基因同时编辑(效率为8.9%)。进一步用编辑细胞进行体细胞核移植生产胚胎,与未编辑细胞生产的胚胎在体细胞融合率、胚胎卵裂率和囊胚率上均无显著差异;测序结果显示,胚胎水平的打靶结果与体细胞一致。综上,利用CRISPR/Cas9技术制备猪MSTN、pAPN、CD163基因同时编辑的PEF和胚胎,为多基因编辑克隆猪的制备奠定基础和提供参考。
中图分类号:
谢浩, 陈指龙, 彭翠婷, 潘颖婷, 齐霖, 赵玉兰, 闭英连, 宋子仪, 唐中林. 猪MSTN、pAPN和CD163基因同步编辑的胚胎制备[J]. 中国农业科技导报, 2025, 27(11): 226-239.
Hao XIE, Zhilong CHEN, Cuiting PENG, Yingting PAN, Lin QI, Yulan ZHAO, Yinglian BI, Ziyi SONG, Zhonglin TANG. Preparation of Embryos with Simultaneous Editing of MSTN, pAPN and CD163 Genes in Pig[J]. Journal of Agricultural Science and Technology, 2025, 27(11): 226-239.
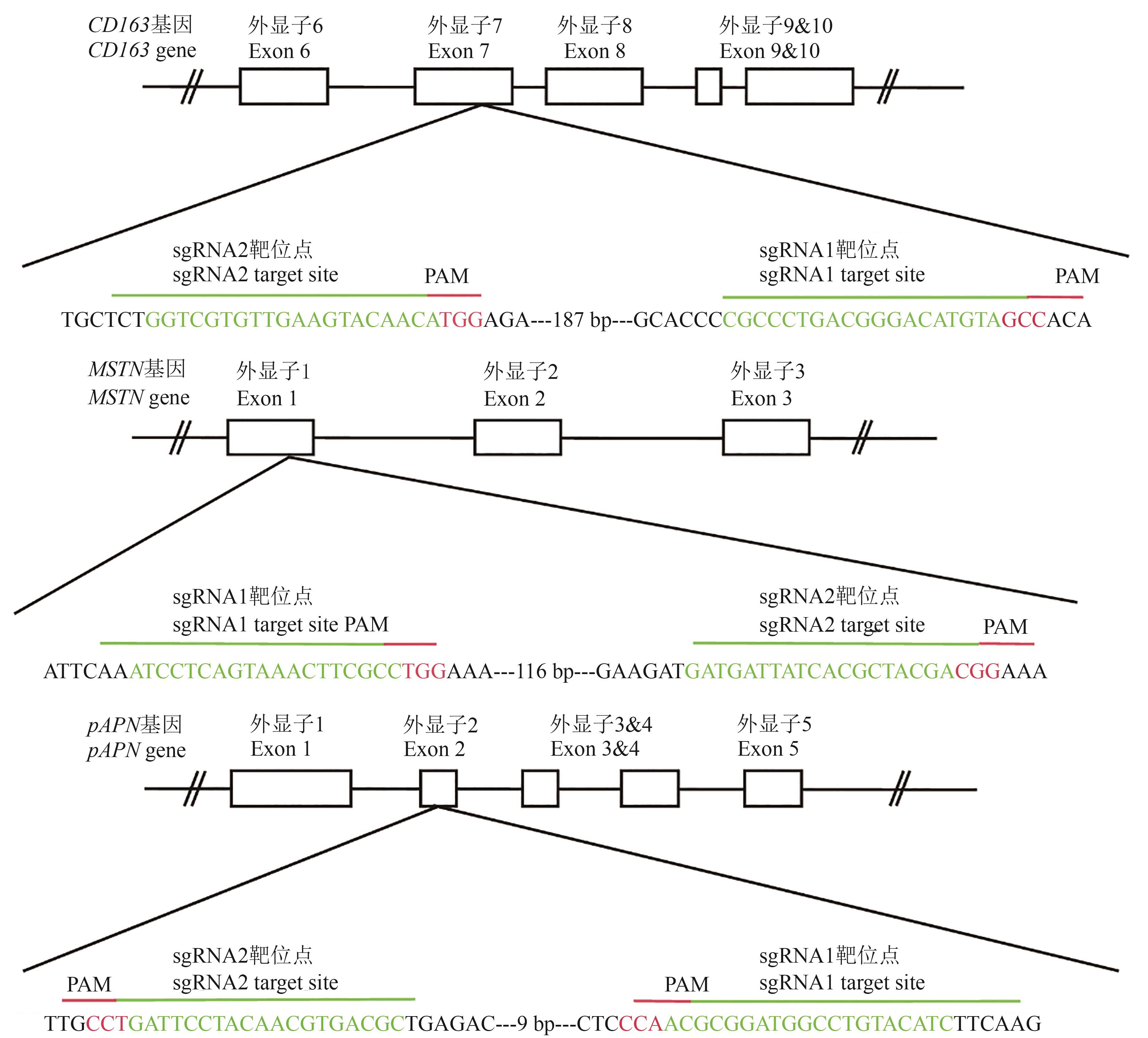
图 1 MSTN、 CD163、pAPN基因的sgRNA位置和序列注:方框为外显子;绿色为sgRNA序列;红色为PAM(protospacer adjacent motif)序列。
Fig. 1 sgRNA positions and sequences of MSTN, CD163 and pAPN genesNote:The box indicates exon; green indicates sgRNA sequence; red indicates PAM sequences.
| 引物名称Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| CD163-sgRNA1 | F: | R: |
| CD163-sgRNA2 | F: | R: |
| MSTN-sgRNA1 | F: | R: |
| MSTN-sgRNA2 | F: | R: |
| pAPN-sgRNA1 | F: | R: |
| pAPN-sgRNA2 | F: | R: |
表1 sgRNA寡聚核苷酸序列
Table 1 Oligo nucleotide sequence of sgRNA
| 引物名称Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| CD163-sgRNA1 | F: | R: |
| CD163-sgRNA2 | F: | R: |
| MSTN-sgRNA1 | F: | R: |
| MSTN-sgRNA2 | F: | R: |
| pAPN-sgRNA1 | F: | R: |
| pAPN-sgRNA2 | F: | R: |
引物名称 Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| 上游Forward | 下游Reverse | |
| CD163-ko | ATGGGTTCCAGAAGGCAAAG | CCATTCACCAAGCGGATTT |
| pAPN-ko | TGTCTGAGCCCTGGTTAATTT | TTGAGCTTCTTGCTATGGATG |
| MSTN-ko | TGAATGAGAACAGCGAGCAA | ATGCCTATTTCAGACAACCAAC |
| ucs-homo | acaaatggctctagaggtaccGAGGGCCTATTTCCCATGATTC | atcatgggaaataggccctcGCACCGACTCGGTGCCAC |
| uas-homo | GAGGGCCTATTTCCCATGATTC | gtaagttatgtaacgggtaccGCACCGACTCGGTGCCAC |
| MSTNsg1-OT-1 | TGCAAAGCTGGACCCACAAAG | AGCTGCAGATGCTCACCTG |
| MSTNsg1-OT-2 | GTAGTGTAGGCCAGCAGCTCTA | ATCTCCAGAGAATCAGGCTGA |
| MSTNsg1-OT-3 | CTTTTCCGCCAAAGCTGTTT | GTTCTCAGACCATGACTATGG |
| CD163sg1-OT-1 | GGTCTCAAACGTCTCCCCT | ACCCGCTCTCCCCCTTCTC |
| CD163sg1-OT-2 | GTTCCCAGGACTGGAGAGG | GTGTCCCTGCTCCCCAGG |
| CD163sg1-OT-3 | TTCCTGACCACCCCACCC | GCCACTGCAACACCAGATCT |
| pAPNsg1-OT-1 | GTGATTTCCCGAAGCCTGTT | GGCTGGGGGTTCCTTCCT |
| pAPNsg1-OT-2 | AGGCTTCCGGAAAATTAAGCTA | GGAGCAGATTAACAGAGACC |
| pAPNsg1-OT-3 | GGAGCAAGTGTGGTACCATG | TTGGAAGATTCACAACTGTAGA |
表2 引物序列
Table 2 Primer sequence
引物名称 Prime name | 引物序列Primer sequence (5’-3’) | |
|---|---|---|
| 上游Forward | 下游Reverse | |
| CD163-ko | ATGGGTTCCAGAAGGCAAAG | CCATTCACCAAGCGGATTT |
| pAPN-ko | TGTCTGAGCCCTGGTTAATTT | TTGAGCTTCTTGCTATGGATG |
| MSTN-ko | TGAATGAGAACAGCGAGCAA | ATGCCTATTTCAGACAACCAAC |
| ucs-homo | acaaatggctctagaggtaccGAGGGCCTATTTCCCATGATTC | atcatgggaaataggccctcGCACCGACTCGGTGCCAC |
| uas-homo | GAGGGCCTATTTCCCATGATTC | gtaagttatgtaacgggtaccGCACCGACTCGGTGCCAC |
| MSTNsg1-OT-1 | TGCAAAGCTGGACCCACAAAG | AGCTGCAGATGCTCACCTG |
| MSTNsg1-OT-2 | GTAGTGTAGGCCAGCAGCTCTA | ATCTCCAGAGAATCAGGCTGA |
| MSTNsg1-OT-3 | CTTTTCCGCCAAAGCTGTTT | GTTCTCAGACCATGACTATGG |
| CD163sg1-OT-1 | GGTCTCAAACGTCTCCCCT | ACCCGCTCTCCCCCTTCTC |
| CD163sg1-OT-2 | GTTCCCAGGACTGGAGAGG | GTGTCCCTGCTCCCCAGG |
| CD163sg1-OT-3 | TTCCTGACCACCCCACCC | GCCACTGCAACACCAGATCT |
| pAPNsg1-OT-1 | GTGATTTCCCGAAGCCTGTT | GGCTGGGGGTTCCTTCCT |
| pAPNsg1-OT-2 | AGGCTTCCGGAAAATTAAGCTA | GGAGCAGATTAACAGAGACC |
| pAPNsg1-OT-3 | GGAGCAAGTGTGGTACCATG | TTGGAAGATTCACAACTGTAGA |
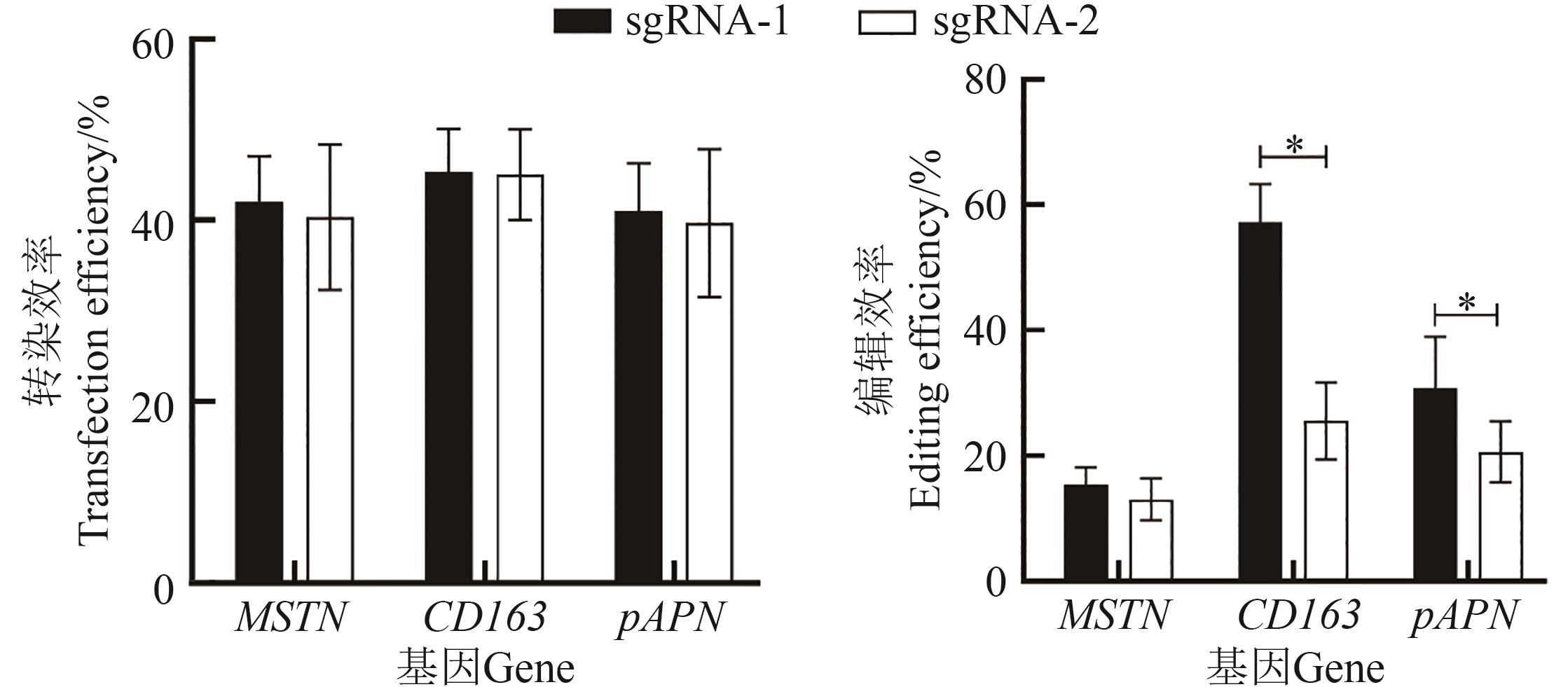
图3 MSTN、CD163和pAPN基因不同sgRNA转染及编辑效率注:*表示在P<0.05水平差异显著。
Fig. 3 Transfection and editing efficiency of different sgRNAs targeting MSTN, CD163 and pAPN genesNote:* indicates significant difference at P<0.05 level.
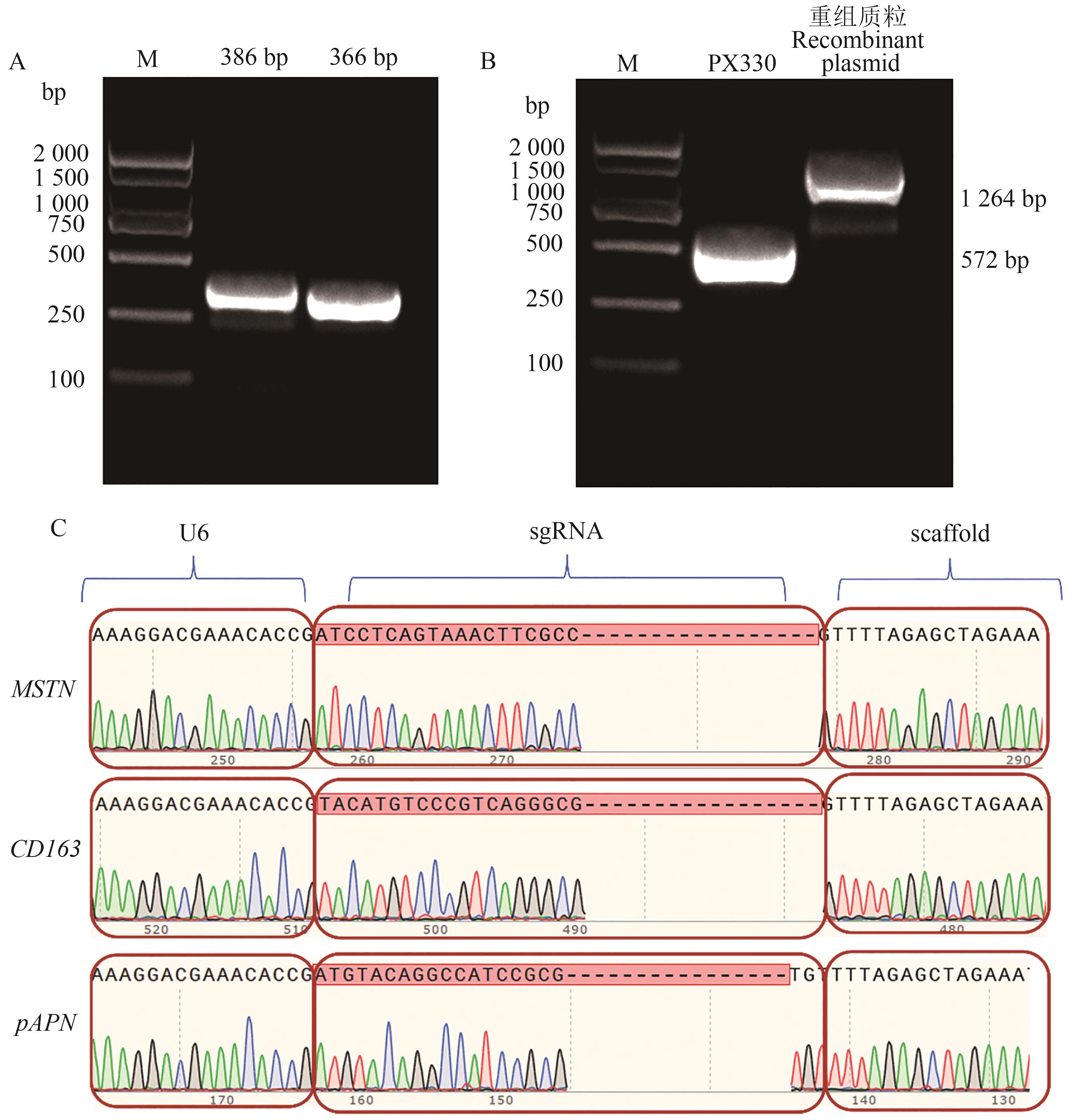
图4 MSTN、CD163和pAPN多基因编辑质粒的鉴定A:片段扩增产物;B重组质粒PCR; C:多基因编辑质粒的测序。M—2 000 bp DNA Marker
Fig. 4 Identification of multi-gene editing plasmid with MSTN, CD163 and pAPN genesA: Products of amplified fragments; B: PCR of recombinant plasmid; C: Ssequencing of multi-gene editing plasmid. M—2 000 bp DNA Marker

图6 三基因敲除细胞缺失序列注:1#~45#为不同单克隆;绿色字体表示原序列;红色字体表示替换;橙色字体表示插入;红色线段表示缺失。
Fig. 6 Deletion sequences of 3 gene in knock out cellNote:1#~45# are different monoclones; the green font indicates the original sequence; the red font indicates the substitutions; the orange font indicates the insertions; the red dashes indicates the deletions.
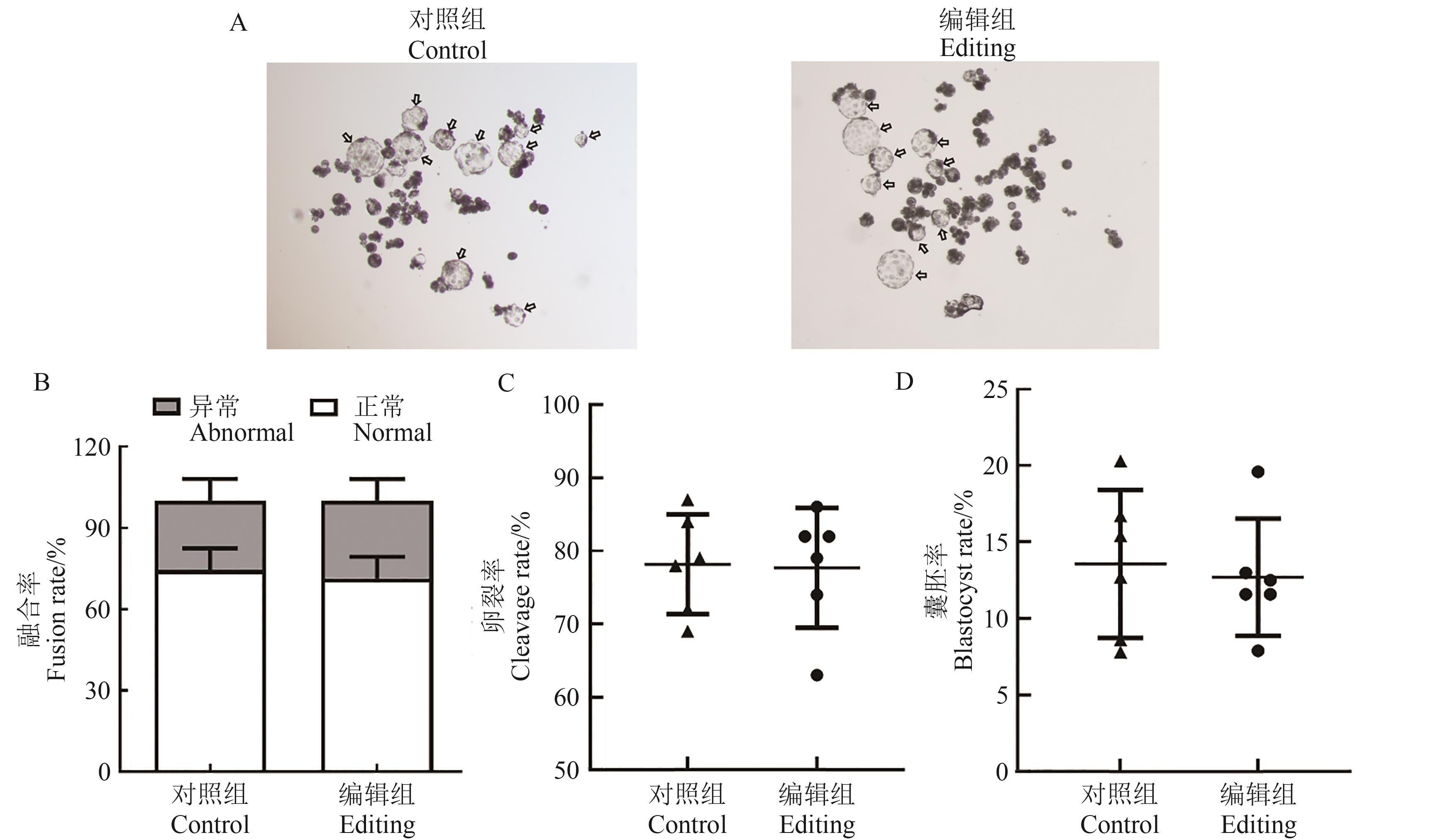
图7 胚胎水平MSTN、CD163、pAPN多基因编辑的验证A:各组囊胚代表图,箭头所示为囊胚;B:体细胞融合率;C:胚胎卵裂率;D:各组囊胚形成率
Fig. 7 Verification of multi-gene editing of MSTN, CD163 and pAPN at embryonic levelA: Representative images of blastocysts in each group, the arrows indicate blastocysts; B: Somatic cell fusion rate; C: Embryo cleavage rate; D: Blastocyst formation rate in each group
| [1] | LIU W, LI L, JIANG J, et al.. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics [J]. Precis. Clin. Med., 2021, 4(3): 179-191. |
| [2] | CHOJNACKA-PUCHTA L, SAWICKA D. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications [J]. J. Appl. Genet., 2020, 61(2): 221-229. |
| [3] | WHYTE J J, PRATHER R S. Genetic modifications of pigs for medicine and agriculture [J]. Mol. Reprod. Dev., 2011, 78(10-11): 879-891. |
| [4] | BIBIKOVA M, BEUMER K, TRAUTMAN J K, et al.. Enhancing gene targeting with designed zinc finger nucleases [J/OL]. Science, 2003, 300(5620): 764 [2025-03-20]. . |
| [5] | MILLER J C, TAN S, QIAO G, et al.. A TALE nuclease architecture for efficient genome editing [J]. Nat. Biotechnol., 2011, 29(2): 143-148. |
| [6] | HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and archaea [J]. Science, 2010, 327(5962): 167-170. |
| [7] | CHEN K, WANG Y, ZHANG R, et al.. CRISPR/cas genome editing and precision plant breeding in agriculture [J]. Annu. Rev. Plant Biol., 2019, 70: 667-697. |
| [8] | CHEN J, WANG H, BAI J, et al.. Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163 [J]. Int. J. Biol. Sci., 2019, 15(2): 481-492. |
| [9] | RAN F A, HSU P D, WRIGHT J, et al.. Genome engineering using the CRISPR-Cas9 system [J]. Nat. Protoc., 2013, 8(11): 2281-2308. |
| [10] | FAN Z, LIU Z, XU K, et al.. Long-term, multidomain analyses to identify the breed and allelic effects in MSTN-edited pigs to overcome lameness and sustainably improve nutritional meat production [J]. Sci. China Life Sci., 2022, 65(2): 362-375. |
| [11] | CHEN J, WANG H, BAI J, et al.. Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163 [J]. Int. J. Biol. Sci., 2019, 15(2): 481-492. |
| [12] | LUO L, WANG S, ZHU L, et al.. Aminopeptidase N-null neonatal piglets are protected from transmissible gastroenteritis virus but not porcine epidemic diarrhea virus [J/OL]. Sci. Rep., 2019, 9(1): 13186 [2025-03-20]. . |
| [13] | KABADI A M, OUSTEROUT D G, HILTON I B, et al.. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector [J/OL]. Nucleic Acids Res., 2014, 42(19): e147 [2025-03-20]. . |
| [14] | DONG F, XIE K, CHEN Y, et al.. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells [J]. Biochem. Biophys. Res. Commun., 2017, 482(4): 889-895. |
| [15] | ZHANG J Q, GUO J X, WU X J, et al.. Optimization of sgRNA expression strategy to generate multiplex gene-edited pigs [J]. Zool. Res., 2022, 43(6): 1005-1008. |
| [16] | HAURWITZ R E, STERNBERG S H, DOUDNA J A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA [J]. EMBO J., 2012, 31(12): 2824-2832. |
| [17] | KISHIMOTO T, NISHIMURA K, MORISHITA K, et al.. An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors [J/OL]. J. Biol. Eng., 2024, 18(1): 9 [2025-03-20]. . |
| [18] | GAO Y, ZHAO Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing [J]. J. Integr. Plant Biol., 2014, 56(4): 343-349. |
| [19] | WANG H, SHEN L, CHEN J, et al.. Deletion of CD163 exon 7 confers resistance to highly pathogenic porcine reproductive and respiratory viruses on pigs [J]. Int. J. Biol. Sci., 2019, 15(9): 1993-2005. |
| [20] | CALVERT J G, SLADE D E, SHIELDS S L, et al.. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses [J]. J. Virol., 2007, 81(14): 7371-7379. |
| [21] | PATTON J B, ROWLAND R R, YOO D, et al.. Modulation of CD163 receptor expression and replication of porcine reproductive and respiratory syndrome virus in porcine macrophages [J]. Virus Res., 2009, 140(1-2): 161-171. |
| [22] | VAN GORP H, VAN BREEDAM W, VAN DOORSSELAERE J, et al.. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus [J]. J. Virol., 2010, 84(6): 3101-3105. |
| [23] | JI C M, WANG B, ZHOU J, et al.. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into vero or porcine small intestine epithelial cells [J]. Virology, 2018, 517: 16-23. |
| [24] | ZHU X, LIU S, WANG X, et al.. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection [J/OL]. Emerg. Microbes Infect., 2018, 7(1): 65 [2025-03-20]. . |
| [25] | WHITWORTH K M, ROWLAND R R R, PETROVAN V, et al.. Resistance to coronavirus infection in amino peptidase N-deficient pigs [J]. Transgenic Res., 2019, 28(1): 21-32. |
| [26] | PENG D W, LI R Q, ZENG W, et al.. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Yi Chuan, 2021, 43(3): 261-270. |
| [27] | YANG Z, VAJTA G, XU Y, et al.. Production of pigs by hand-made cloning using mesenchymal stem cells and fibroblasts [J]. Cell. Reprogram., 2016, 18(4): 256-263. |
| [28] | YOSHIOKA K, SUZUKI C, TANAKA A, et al.. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium [J]. Biol. Reprod., 2002, 66(1): 112-119. |
| [29] | 晏超, 刘永刚, 谢浩, 等. 预扩增qPCR技术检测少量猪早期胚胎细胞基因表达的研究[J]. 畜牧兽医学报, 2024, 55(12):5567-5574. |
| YAN C, LIU Y G, XIE H, et al.. Detection of gene expression in trace cells of early porcine embryo by pre-amplified quantitative PCR [J]. Acta Vet. Zootech. Sin., 2024, 55(12): 5567-5574. | |
| [30] | 刘雯雯, 董发明, 毕延震. 多基因编辑技术的发展及其在畜牧种质创新中的应用[J]. 畜牧兽医学报, 2024, 55(8): 3267-3275. |
| LIU W W, DONG F M, BI Y Z. The development of multi-gene editing technology and its application in agricultural biological germplasm innovation [J]. Acta Vet. Zootech. Sin., 2024, 55(8): 3267-3275. | |
| [31] | 郎楠, 梁洛瑜, 汪军丽, 等. CRISPR-Cas9多基因编辑技术在植物研究中的应用[J]. 分子植物育种, 2023, 21(8): 2665-2670. |
| LANG N, LIANG L Y, WANG J L, et al.. Application of CRISPR-Cas9 enabled multiplex gene editing in plant research [J]. Mol. Plant Breeding, 2023, 21(8): 2665-2670. | |
| [32] | XU K, ZHOU Y, MU Y, et al.. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance [J/OL]. eLife, 2020, 9: e57132 [2025-03-20]. . |
| [33] | SAKUMA T, NISHIKAWA A, KUME S, et al.. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system [J/OL]. Sci. Rep., 2014, 4: 5400 [2025-03-20]. . |
| [34] | VAD-NIELSEN J, LIN L, BOLUND L, et al.. Golden gate assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes [J]. Cell. Mol. Life Sci., 2016, 73(22): 4315-4325. |
| [35] | MA X, ZHANG Q, ZHU Q, et al.. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants [J]. Mol. Plant, 2015, 8(8): 1274-1284. |
| [36] | SONG R, WANG Y, ZHENG Q, et al.. One-step base editing in multiple genes by direct embryo injection for pig trait improvement [J]. Sci. China Life Sci., 2022, 65(4): 739-752. |
| [37] | REN J, HAI T, CHEN Y, et al.. Improve meat production and virus resistance by simultaneously editing multiple genes in livestock using Cas12i (Max) [J]. Sci. China Life Sci., 2024, 67(3): 555-564. |
| [38] | ETZERODT A, KJOLBY M, NIELSEN M J, et al.. Plasma clearance of hemoglobin and haptoglobin in mice and effect of CD163 gene targeting disruption [J]. Antioxid. Redox Signal., 2013, 18(17): 2254-2263. |
| [39] | ZHANG Z, BAXTER A E, REN D, et al.. Efficient engineering of human and mouse primary cells using peptide-assisted genome editing [J]. Nat. Biotechnol., 2024, 42(2): 305-315. |
| [40] | XIE K, MINKENBERG B, YANG Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system [J]. Proc. Natl. Acad. Sci. USA, 2015, 112(11): 3570-3575. |
| [41] | ZHANG D, ZHANG H, LI T, et al.. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases [J/OL]. Genome Biol., 2017, 18(1): 191 [2025-03-20]. . |
| [42] | KONSTANTAKOS V, NENTIDIS A, KRITHARA A, et al.. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning [J]. Nucleic Acids Res., 2022, 50(7): 3616-3637. |
| [43] | XU X, DUAN D, CHEN S J. CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment [J/OL]. Sci. Rep., 2017, 7(1): 143 [2025-03-20]. . |
| [44] | DOENCH J G, FUSI N, SULLENDER M, et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 [J]. Nat. Biotechnol., 2016, 34(2): 184-191. |
| [45] | HAN H A, PANG J K S, B-SSOH. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing [J]. J. Mol. Med. (Berl), 2020, 98(5): 615-632. |
| [46] | GUO C, MA X, GAO F, et al.. Off-target effects in CRISPR/Cas9 gene editing [J/OL]. Front. Bioeng. Biotechnol., 2023, 11: 1143157 [2025-03-20]. . |
| [47] | SCHMID-BURGK J L, GAO L, LI D, et al.. Highly parallel profiling of Cas9 variant specificity [J]. Mol. Cell, 2020, 78(4): 794-800.e8. |
| [48] | SCHUSTERBAUER V, FISCHER J E, GANGL S, et al.. Whole genome sequencing analysis of effects of CRISPR/Cas9 in Komagataella phaffii: a budding yeast in distress [J/OL]. J. Fungi (Basel), 2022, 8(10): 992 [2025-03-20]. . |
| [49] | H-HTSAI, H-JKAO, KUO M W, et al.. Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing [J/OL]. Nat. Commun., 2023, 14(1): 5183 [2025-03-20]. . |
| [1] | 张文婷, 李阳, 裘实, 路光明, 郭冬姝, 张保龙, 王金彦. 基于CRISPR/Cas9基因编辑技术研究Badh2基因对稻米品质的影响[J]. 中国农业科技导报, 2025, 27(5): 39-48. |
| [2] | 张才用, 陈指龙, 谢浩, 彭翠婷, 晏超, 赵玉兰, 齐霖, 周磊, 唐中林. 提高猪体细胞克隆胚胎发育率的研究进展[J]. 中国农业科技导报, 2024, 26(12): 201-209. |
| [3] | 王立斌, 王强龙, 潘阳阳, 赵天, 丁天翊, 崔燕, 余四九. 牦牛卵母细胞体外成熟培养及孤雌发育研究[J]. 中国农业科技导报, 2023, 25(10): 84-90. |
| [4] | 谢春嫡, 郭肖蓉, 张煦栋, 周荣, 李奎. 基于CRISPR/Cas9技术建立IGF2R基因修饰PK-15细胞[J]. 中国农业科技导报, 2022, 24(10): 200-207. |
| [5] | 庄重, 赵龙, 白皓, 毕瑜林, 黄应权, 陈国宏, 常国斌. CRISPR/Cas9技术在家禽育种方面的应用[J]. 中国农业科技导报, 2022, 24(1): 14-23. |
| [6] | 廖嘉明, §, 李春梅§, 张石虎, 李布野, 欧阳昆唏, 陈晓阳. CRISPR/Cas9基因编辑技术的发展及其在植物中的应用[J]. 中国农业科技导报, 2021, 23(12): 20-28. |
| [7] | 刘芊萩,陈强,雷安民*. 微流控技术在解决猪体外受精中多精入卵问题中的应用进展[J]. 中国农业科技导报, 2021, 23(1): 66-72. |
| [8] | 辛红佳,李鹏程,滕守振,李圣彦,汪海,郎志宏*. 拟南芥SWEET1/2/3基因突变体构建及功能鉴定[J]. 中国农业科技导报, 2020, 22(2): 39-49. |
| [9] | 刘杜娟,黄火清*,苏小运* . 低纤维素酶背景里氏木霉菌株的构建和应用[J]. 中国农业科技导报, 2020, 22(12): 50-57. |
| [10] | 胡婉彬,李家祥,段立柱,刘敏博,常亚青,卢金一,焦杨鹏,湛垚垚*. 两种海水酸化模式对马粪海胆(Hemicentrotus pulcherrimus)胚胎早期发育的影响[J]. 中国农业科技导报, 2016, 18(3): 177-183. |
| [11] | 王成龙1,2,周美亮2,董雪妮1,2,唐益雄2,邵继荣1*,吴燕民2*. 紫花苜蓿两种再生体系的优化及比较[J]. , 2015, 17(4): 53-61. |
| [12] | 李喜和1,2. 家畜繁育生物技术研究开发与产业化推广应用[J]. , 2013, 15(3): 64-71. |
| [13] | Hunter R H F1,李喜和2,3*. 采用卵子-胚胎移植技术分析研究家畜受精和早期发育[J]. , 2013, 15(1): 65-70. |
| [14] | 张鑫,陈苏仁,栗雪冰,刘东军,仓明. 牛早期胚胎发育中差异基因的表达研究[J]. , 2009, 11(6): 50-54. |
| [15] | 梁秋菊1,2,王崇英1,范云六2,张春义2. 质体发育分化对植物胚胎发育的影响[J]. , 2009, 11(5): 6-11. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||